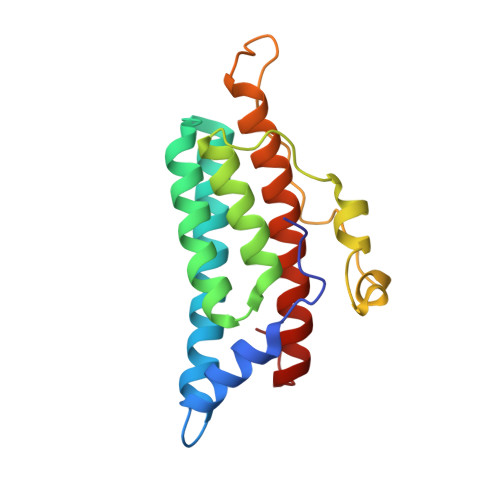
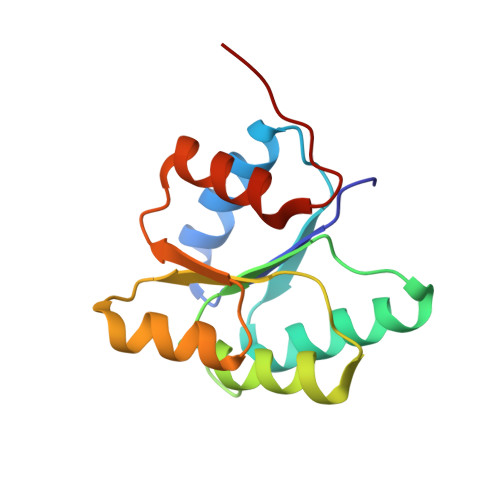
Crystal structure of a complex between the phosphorelay protein YPD1 and the response regulator domain of SLN1 bound to a phosphoryl analog
Zhao, X., Copeland, D.M., Soares, A.S., West, A.H.(2008) J Mol Biology 375: 1141-1151
- PubMed: 18076904
- DOI: https://doi.org/10.1016/j.jmb.2007.11.045
- Primary Citation of Related Structures:
2R25 - PubMed Abstract:
The crystal structure of the yeast SLN1 response regulator (RR) domain bound to both a phosphoryl analog [beryllium fluoride (BeF(3)(-))] and Mg(2+), in complex with its downstream phosphorelay signaling partner YPD1, has been determined at a resolution of 1.70 A. Comparisons between the BeF(3)(-)-activated complex and the unliganded (or apo) complex determined previously reveal modest but important differences. The SLN1-R1 x Mg(2+) x BeF(3)(-) structure from the complex provides evidence for the first time that the mechanism of phosphorylation-induced activation is highly conserved between bacterial RR domains and this example from a eukaryotic organism. Residues in and around the active site undergo slight rearrangements in order to form bonds with the essential divalent cation and fluorine atoms of BeF(3)(-). Two conserved switch-like residues (Thr1173 and Phe1192) occupy distinctly different positions in the apo versus BeF(3)(-)-bound structures, consistent with the "Y-T" coupling mechanism proposed for the activation of CheY and other bacterial RRs. Several loop regions and the alpha 4-beta 5-alpha 5 surface of the SLN1-R1 domain undergo subtle conformational changes ( approximately 1-3 A displacements relative to the apo structure) that lead to significant changes in terms of contacts that are formed with YPD1. Detailed structural comparisons of protein-protein interactions in the apo and BeF(3)(-)-bound complexes suggest at least a two-state equilibrium model for the formation of a transient encounter complex, in which phosphorylation of the RR promotes the formation of a phosphotransfer-competent complex. In the BeF(3)(-)-activated complex, the position of His64 from YPD1 needs to be within ideal distance of and in near-linear geometry with Asp1144 from the SLN1-R1 domain for phosphotransfer to occur. The ground-state structure presented here suggests that phosphoryl transfer will likely proceed through an associative mechanism involving the formation of a pentacoordinate phosphorus intermediate.
- Department of Chemistry and Biochemistry, University of Oklahoma, 620 Parrington Oval, Norman, OK 73019, USA.
Organizational Affiliation: