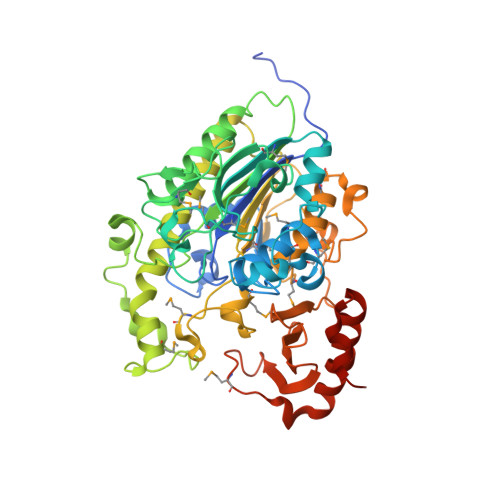
X-ray crystal structure of the putative sulfatase yidJ from Bacteroides fragilis.
Vorobiev, S.M., Abashidze, M., Seetharaman, J., Wang, D., Cunningham, K., Maglaqui, M., Owens, L., Xiao, R., Acton, T.B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.