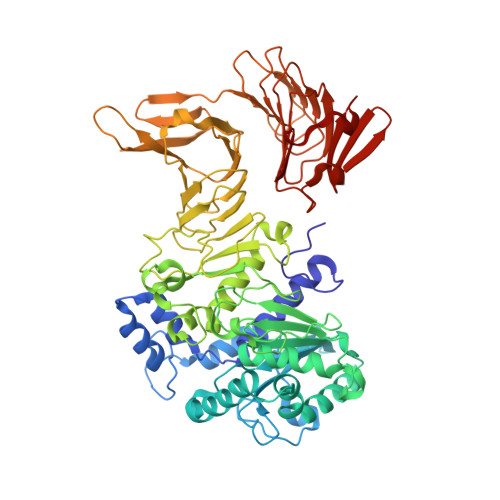
A calcium-gated lid and a large beta-roll sandwich are revealed by the crystal structure of extracellular lipase from Serratia marcescens.
Meier, R., Drepper, T., Svensson, V., Jaeger, K.E., Baumann, U.(2007) J Biological Chem 282: 31477-31483
- PubMed: 17728256
- DOI: https://doi.org/10.1074/jbc.M704942200
- Primary Citation of Related Structures:
2QUA, 2QUB - PubMed Abstract:
Lipase LipA from Serratia marcescens is a 613-amino acid enzyme belonging to family I.3 of lipolytic enzymes that has an important biotechnological application in the production of a chiral precursor for the coronary vasodilator diltiazem. Like other family I.3 lipases, LipA is secreted by Gram-negative bacteria via a type I secretion system and possesses 13 copies of a calcium binding tandem repeat motif, GGXGXDXUX (U, hydrophobic amino acids), in the C-terminal part of the polypeptide chain. The 1.8-A crystal structure of LipA reveals a close relation to eukaryotic lipases, whereas family I.1 and I.2 enzymes appear to be more distantly related. Interestingly, the structure shows for the N-terminal lipase domain a variation on the canonical alpha/beta hydrolase fold in an open conformation, where the putative lid helix is anchored by a Ca(2+) ion essential for activity. Another novel feature observed in this lipase structure is the presence of a helical hairpin additional to the putative lid helix that exposes a hydrophobic surface to the aqueous medium and might function as an additional lid. The tandem repeats form two separated parallel beta-roll domains that pack tightly against each other. Variations of the consensus sequence of the tandem repeats within the second beta-roll result in an asymmetric Ca(2+) binding on only one side of the roll. The analysis of the properties of the beta-roll domains suggests an intramolecular chaperone function.
- Department of Chemistry and Biochemistry, University Bern Freiestrasse 3, CH-3012 Bern, Switzerland.
Organizational Affiliation: