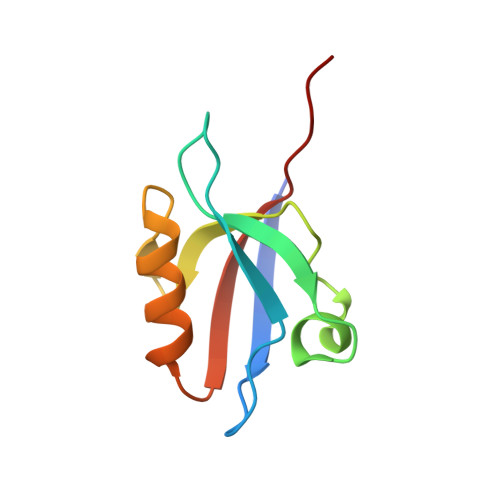
Crystal structure of the 11th PDZ domain of MPDZ (MUPP1).
Papagrigoriou, E., Salah, E., Phillips, C., Savitsky, P., Boisguerin, P., Oschkinat, H., Gileadi, C., Yang, X., Elkins, J.M., Ugochukwu, E., Bunkoczi, G., Uppenberg, J., Doyle, D.To be published.