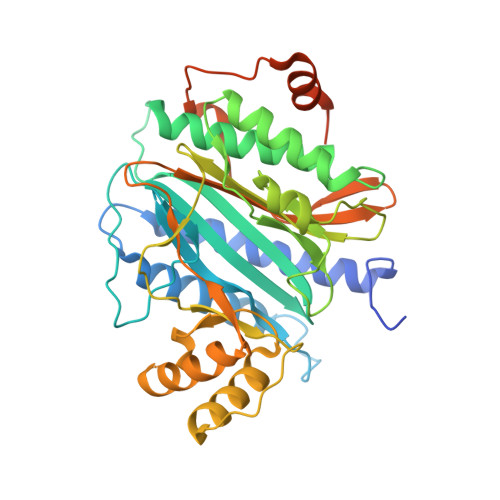
The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions.
Kowalinski, E., Bange, G., Bradatsch, B., Hurt, E., Wild, K., Sinning, I.(2007) FEBS Lett 581: 4450-4454
- PubMed: 17765895
- DOI: https://doi.org/10.1016/j.febslet.2007.08.024
- Primary Citation of Related Structures:
2Q8K - PubMed Abstract:
The ErbB-3 receptor binding protein (Ebp1) is a member of the proliferation-associated 2G4 (PA2G4) family implicated in regulation of cell growth and differentiation. Here, we report the crystal structure of the human Ebp1 at 1.6 A resolution. The protein has the conserved pita bread fold of methionine aminopeptidases, but without the characteristic enzymatic activity. Moreover, Ebp1 is known to interact with a number of proteins and RNAs involved in either transcription regulation or translation control. The structure provides insights in how Ebp1 discriminates between its different interaction partners.
- Heidelberg University Biochemistry Center (BZH), INF 328, D-69120 Heidelberg, Germany.
Organizational Affiliation: