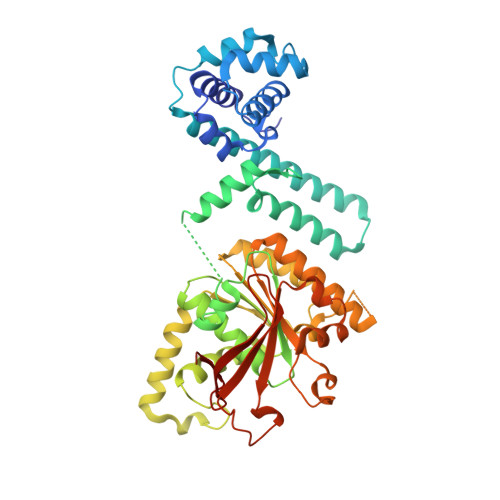
The crystal structure of the Thermus aquaticus DnaB helicase monomer.
Bailey, S., Eliason, W.K., Steitz, T.A.(2007) Nucleic Acids Res 35: 4728-4736
- PubMed: 17606462
- DOI: https://doi.org/10.1093/nar/gkm507
- Primary Citation of Related Structures:
2Q6T - PubMed Abstract:
The ring-shaped hexameric DnaB helicase unwinds duplex DNA at the replication fork of eubacteria. We have solved the crystal structure of the full-length Thermus aquaticus DnaB monomer, or possibly dimer, at 2.9 A resolution. DnaB is a highly flexible two domain protein. The C-terminal domain exhibits a RecA-like core fold and contains all the conserved sequence motifs that are characteristic of the DnaB helicase family. The N-terminal domain contains an additional helical hairpin that makes it larger than previously appreciated. Several DnaB mutations that modulate its interaction with primase are found in this hairpin. The similarity in the fold of the DnaB N-terminal domain with that of the C-terminal helicase-binding domain (HBD) of the DnaG primase also includes this hairpin. Comparison of hexameric homology models of DnaB with the structure of the papillomavirus E1 helicase suggests the two helicases may function through different mechanisms despite their sharing a common ancestor.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520, USA.
Organizational Affiliation: