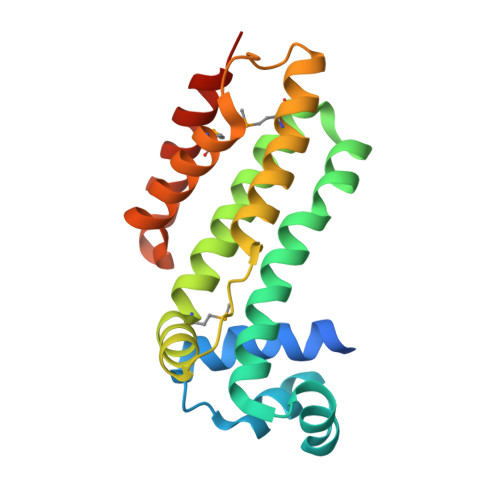
Crystal structure of a putative transcriptional regulator SCO0520 from Streptomyces coelicolor A3(2) reveals an unusual dimer among TetR family proteins.
Filippova, E.V., Chruszcz, M., Cymborowski, M., Gu, J., Savchenko, A., Edwards, A., Minor, W.(2011) J Struct Funct Genomics 12: 149-157
- PubMed: 21625866
- DOI: https://doi.org/10.1007/s10969-011-9112-4
- Primary Citation of Related Structures:
2Q24 - PubMed Abstract:
A structure of the apo-form of the putative transcriptional regulator SCO0520 from Streptomyces coelicolor A3(2) was determined at 1.8 Å resolution. SCO0520 belongs to the TetR family of regulators. In the crystal lattice, the asymmetric unit contains two monomers that form an Ω-shaped dimer. The distance between the two DNA-recognition domains is much longer than the corresponding distances in the known structures of other TetR family proteins. In addition, the subunits in the dimer have different conformational states, resulting in different relative positions of the DNA-binding and regulatory domains. Similar conformational modifications are observed in other TetR regulators and result from ligand binding. These studies provide information about the flexibility of SCO0520 molecule and its putative biological function.
- Department of Molecular Physiology and Biological Physics, University of Virginia, 1340 Jefferson Park Avenue, Jordan Hall, Room 4223, Charlottesville, VA 22908, USA.
Organizational Affiliation: