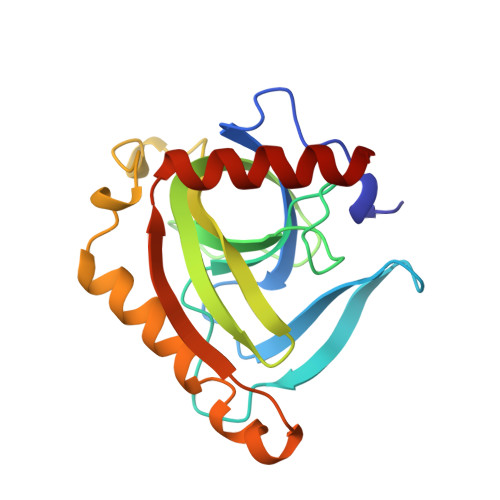
Crystal structure of inorganic pyrophosphatase from Thermus thermophilus.
Teplyakov, A., Obmolova, G., Wilson, K.S., Ishii, K., Kaji, H., Samejima, T., Kuranova, I.(1994) Protein Sci 3: 1098-1107
- PubMed: 7920256
- DOI: https://doi.org/10.1002/pro.5560030713
- Primary Citation of Related Structures:
2PRD - PubMed Abstract:
The 3-dimensional structure of inorganic pyrophosphatase from Thermus thermophilus (T-PPase) has been determined by X-ray diffraction at 2.0 A resolution and refined to R = 15.3%. The structure consists of an antiparallel closed beta-sheet and 2 alpha-helices and resembles that of the yeast enzyme in spite of the large difference in size (174 and 286 residues, respectively), little sequence similarity beyond the active center (about 20%), and different oligomeric organization (hexameric and dimeric, respectively). The similarity of the polypeptide folding in the 2 PPases provides a very strong argument in favor of an evolutionary relationship between the yeast and bacterial enzymes. The same Greek-key topology of the 5-stranded beta-barrel was found in the OB-fold proteins, the bacteriophage gene-5 DNA-binding protein, toxic-shock syndrome toxin-1, and the major cold-shock protein of Bacillus subtilis. Moreover, all known nucleotide-binding sites in these proteins are located on the same side of the beta-barrel as the active center in T-PPase. Analysis of the active center of T-PPase revealed 17 residues of potential functional importance, 16 of which are strictly conserved in all sequences of soluble PPases. Their possible role in the catalytic mechanism is discussed on the basis of the present crystal structure and with respect to site-directed mutagenesis studies on the Escherichia coli enzyme. The observed oligomeric organization of T-PPase allows us to suggest a possible mechanism for the allosteric regulation of hexameric PPases.
- European Molecular Biology Laboratory, Hamburg, Germany.
Organizational Affiliation: