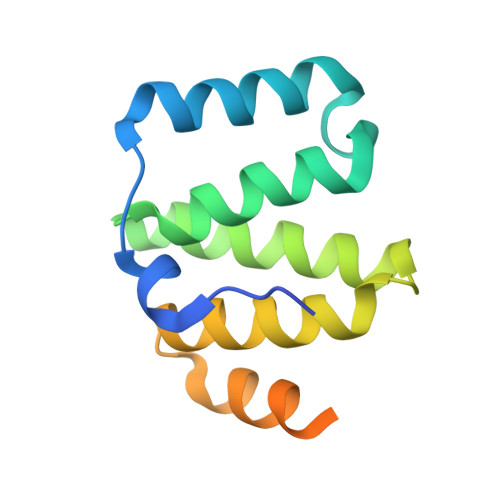
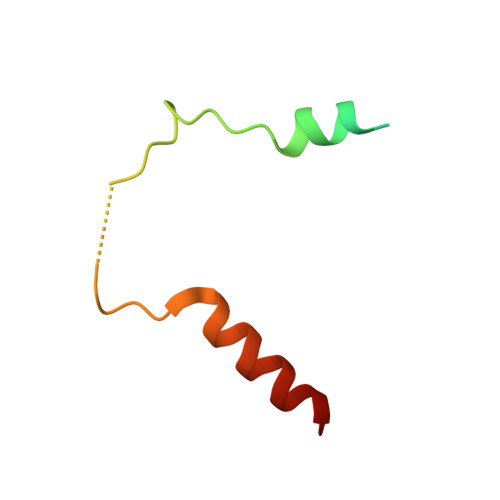
Structural basis for recruitment of mitochondrial fission complexes by Fis1.
Zhang, Y., Chan, D.C.(2007) Proc Natl Acad Sci U S A 104: 18526-18530
- PubMed: 17998537
- DOI: https://doi.org/10.1073/pnas.0706441104
- Primary Citation of Related Structures:
2PQN, 2PQR - PubMed Abstract:
Mitochondrial fission controls mitochondrial shape and physiology, including mitochondrial remodeling in apoptosis. During assembly of the yeast mitochondrial fission complex, the outer membrane protein Fis1 recruits the dynamin-related GTPase Dnm1 to mitochondria. Fis1 contains a tetratricopeptide repeat (TPR) domain and interacts with Dnm1 via the molecular adaptors Mdv1 and Caf4. By using crystallographic analysis of adaptor-Fis1 complexes, we show that these adaptors use two helices to bind to both the concave and convex surfaces of the Fis1 TPR domain. Fis1 therefore contains two interaction interfaces, a binding mode that, to our knowledge, has not been observed previously for TPR domains. Genetic and biochemical studies indicate that both binding interfaces are important for binding of Mdv1 and Caf4 to Fis1 and for mitochondrial fission activity in vivo. Our results reveal how Fis1 recruits the mitochondrial fission complex and will facilitate efforts to manipulate mitochondrial fission.
- Division of Biology, California Institute of Technology, 1200 East California Boulevard, MC 114-96, Pasadena, CA 91125, USA.
Organizational Affiliation: