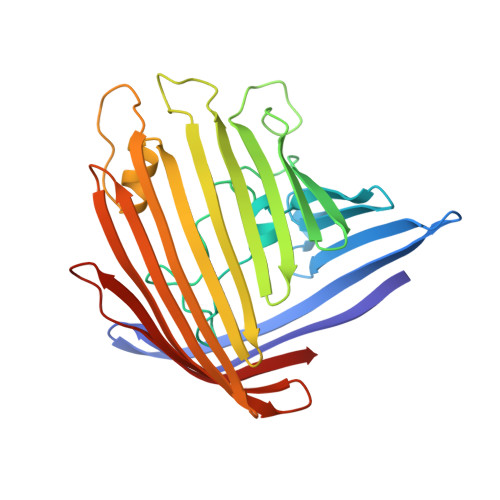
Structure of porin refined at 1.8 A resolution.
Weiss, M.S., Schulz, G.E.(1992) J Mol Biology 227: 493-509
- PubMed: 1328651
- DOI: https://doi.org/10.1016/0022-2836(92)90903-w
- Primary Citation of Related Structures:
2POR - PubMed Abstract:
The crystal structure of porin from Rhodobacter capsulatus has been refined using the simulated annealing method. The final model consists of all 301 amino acid residues well obeying standard geometry, three calcium ions, 274 solvent molecules, three detergent molecules and one unknown ligand modeled as a detergent molecule. The final crystallographic R-factor is 18.6% based on 42,851 independent reflections in the resolution range 10 to 1.8 A. The model is described in detail.
- Institut für Organische Chemie und Biochemie der Universität, Freiburg, Germany.
Organizational Affiliation: