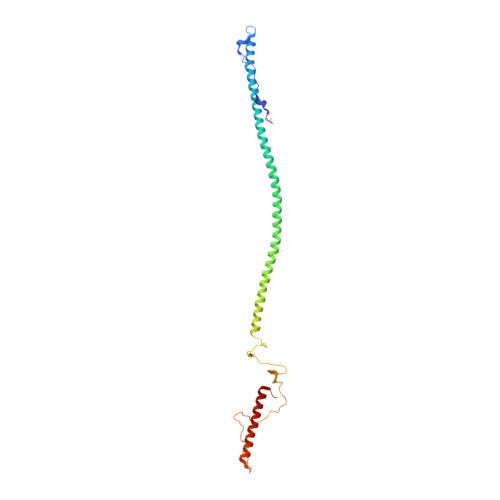
Structure of phage P22 cell envelope-penetrating needle.
Olia, A.S., Casjens, S., Cingolani, G.(2007) Nat Struct Mol Biol 14: 1221-1226
- PubMed: 18059287
- DOI: https://doi.org/10.1038/nsmb1317
- Primary Citation of Related Structures:
2POH - PubMed Abstract:
Bacteriophage P22 infects Salmonella enterica by injecting its genetic material through the cell envelope. During infection, a specialized tail needle, gp26, is injected into the host, likely piercing a hole in the host cell envelope. The 2.1-Å crystal structure of gp26 reveals a 240-Å elongated protein fiber formed by two trimeric coiled-coil domains interrupted by a triple β-helix.The N terminus of gp26 plugs the portal protein channel, retaining the genetic material inside the virion. The C-terminal tip of the fiber exposes β-hairpins with hydrophobic tips similar to those seen in class II fusion peptides. The α-helical core connecting these two functionally polarized tips presents four trimerization octads with consensus sequence IXXLXXXV. The slender conformation of the gp26 fiber minimizes the surface exposed to solvent, which is consistent with the idea that gp26 traverses the cell envelope lipid bilayers.
- Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, 750 East Adams Street, Syracuse, New York 13210, USA.
Organizational Affiliation: