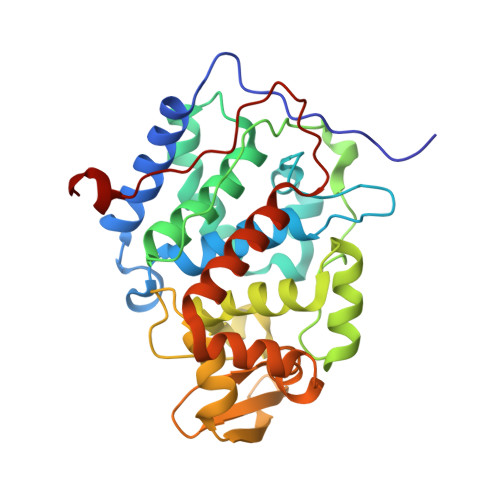
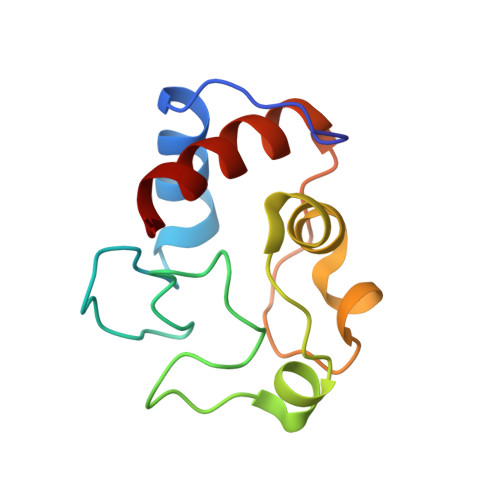
Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c.
Pelletier, H., Kraut, J.(1992) Science 258: 1748-1755
- PubMed: 1334573
- DOI: https://doi.org/10.1126/science.1334573
- Primary Citation of Related Structures:
2PCB, 2PCC - PubMed Abstract:
The crystal structure of a 1:1 complex between yeast cytochrome c peroxidase and yeast iso-1-cytochrome c was determined at 2.3 A resolution. This structure reveals a possible electron transfer pathway unlike any previously proposed for this extensively studied redox pair. The shortest straight line between the two hemes closely follows the peroxidase backbone chain of residues Ala194, Ala193, Gly192, and finally Trp191, the indole ring of which is perpendicular to, and in van der Waals contact with, the peroxidase heme. The crystal structure at 2.8 A of a complex between yeast cytochrome c peroxidase and horse heart cytochrome c was also determined. Although crystals of the two complexes (one with cytochrome c from yeast and the other with cytochrome c from horse) grew under very different conditions and belong to different space groups, the two complex structures are closely similar, suggesting that cytochrome c interacts with its redox partners in a highly specific manner.
- Department of Chemistry, University of California, San Diego, La Jolla 92093-0317.
Organizational Affiliation: