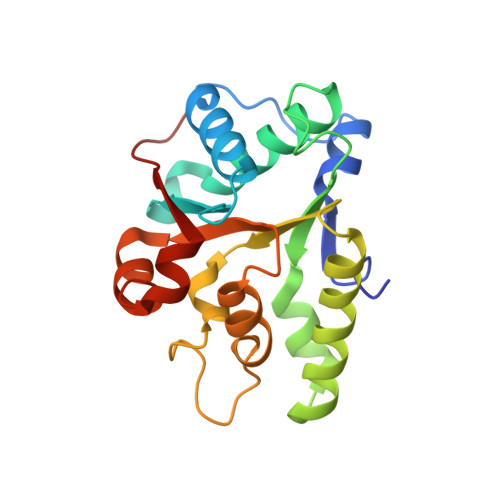
Crystal Structure of the first nucleotide binding domain of chaperone ClpB1, putative, (Pv089580) from Plasmodium Vivax
Wernimont, A.K., Lew, J., Kozieradzki, I., Lin, Y.H., Hassanali, A., Zhao, Y., Arrowsmith, C.H., Edwards, A.M., Weigelt, J., Sundstrom, M., Bochkarev, A., Hui, R., Artz, J.D.To be published.