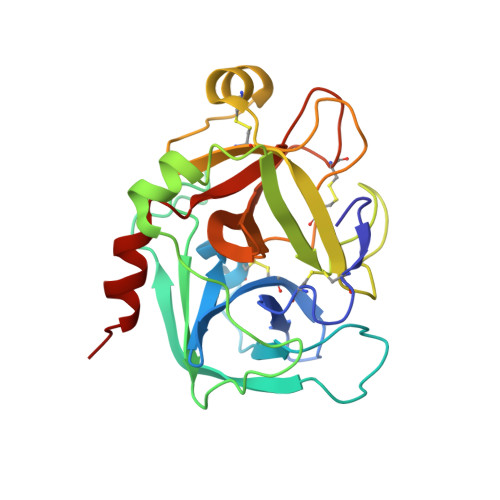
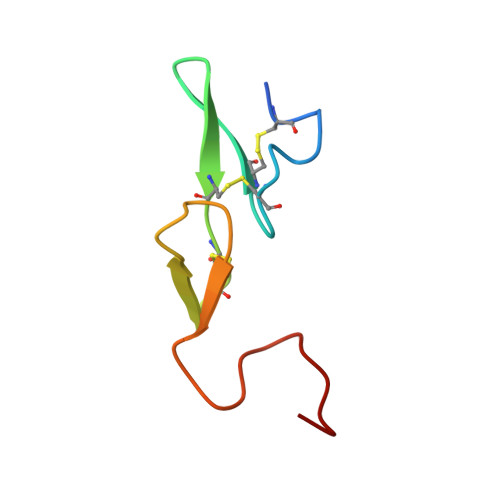
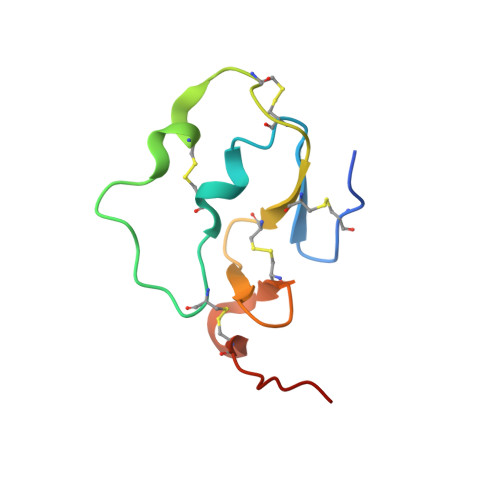
Active and exo-site inhibition of human factor Xa: structure of des-Gla factor Xa inhibited by NAP5, a potent nematode anticoagulant protein from Ancylostoma caninum
Rios-Steiner, J.L., Murakami, M.T., Tulinsky, A., Arni, R.K.(2007) J Mol Biology 371: 774-786
- PubMed: 17588602
- DOI: https://doi.org/10.1016/j.jmb.2007.05.042
- Primary Citation of Related Structures:
2P3F - PubMed Abstract:
Hookworms are hematophagous nematodes capable of growth, development and subsistence in living host systems such as humans and other mammals. Approximately one billion, or one in six, people worldwide are infected by hookworms causing gastrointestinal blood loss and iron deficiency anemia. The hematophagous hookworm Ancylostoma caninum produces a family of small, disulfide-linked protein anticoagulants (75-84 amino acid residues). One of these nematode anticoagulant proteins, NAP5, inhibits the amidolytic activity of factor Xa (fXa) with K(i)=43 pM, and is the most potent natural fXa inhibitor identified thus far. The crystal structure of NAP5 bound at the active site of gamma-carboxyglutamic acid domainless factor Xa (des-fXa) has been determined at 3.1 A resolution, which indicates that Asp189 (fXa, S1 subsite) binds to Arg40 (NAP5, P1 site) in a mode similar to that of the BPTI/trypsin interaction. However, the hydroxyl group of Ser39 of NAP5 additionally forms a hydrogen bond (2.5 A) with His57 NE2 of the catalytic triad, replacing the hydrogen bond of Ser195 OG to the latter in the native structure, resulting in an interaction that has not been observed before. Furthermore, the C-terminal extension of NAP5 surprisingly interacts with the fXa exosite of a symmetry-equivalent molecule forming a short intermolecular beta-strand as observed in the structure of the NAPc2/fXa complex. This indicates that NAP5 can bind to fXa at the active site, or the exosite, and to fX at the exosite. However, unlike NAPc2, NAP5 does not inhibit fVIIa of the fVIIa/TF complex.
- Department of Chemistry, Michigan State University, East Lansing, MI 48824-1322, USA.
Organizational Affiliation: