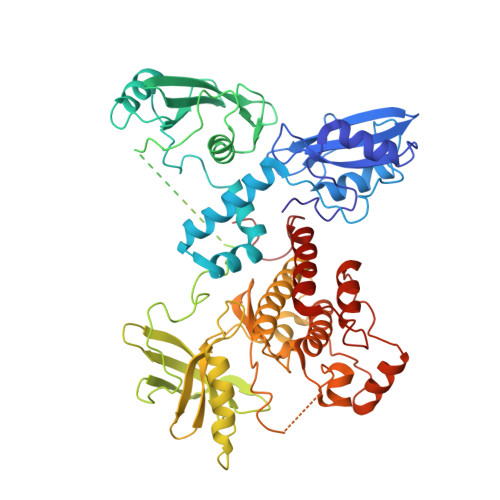
Structural Basis for the Inhibition of Tyrosine Kinase Activity of ZAP-70.
Deindl, S., Kadlecek, T.A., Brdicka, T., Cao, X., Weiss, A., Kuriyan, J.(2007) Cell 129: 735-746
- PubMed: 17512407
- DOI: https://doi.org/10.1016/j.cell.2007.03.039
- Primary Citation of Related Structures:
2OZO - PubMed Abstract:
ZAP-70, a cytoplasmic tyrosine kinase required for T cell antigen receptor signaling, is controlled by a regulatory segment that includes a tandem SH2 unit responsible for binding to immunoreceptor tyrosine-based activation motifs (ITAMs). The crystal structure of autoinhibited ZAP-70 reveals that the inactive kinase domain adopts a conformation similar to that of cyclin-dependent kinases and Src kinases. The autoinhibitory mechanism of ZAP-70 is, however, distinct and involves interactions between the regulatory segment and the hinge region of the kinase domain that reduce its flexibility. Two tyrosine residues in the SH2-kinase linker that activate ZAP-70 when phosphorylated are involved in aromatic-aromatic interactions that connect the linker to the kinase domain. These interactions are inconsistent with ITAM binding, suggesting that destabilization of this autoinhibited ZAP-70 conformation is the first step in kinase activation.
- Department of Molecular and Cell Biology, Department of Chemistry, and Howard Hughes Medical Institute, University of California, Berkeley, CA 94720, USA.
Organizational Affiliation: