Structural basis for converting a general transcription factor into an operon-specific virulence regulator.
Belogurov, G.A., Vassylyeva, M.N., Svetlov, V., Klyuyev, S., Grishin, N.V., Vassylyev, D.G., Artsimovitch, I.(2007) Mol Cell 26: 117-129
- PubMed: 17434131
- DOI: https://doi.org/10.1016/j.molcel.2007.02.021
- Primary Citation of Related Structures:
2OUG - PubMed Abstract:
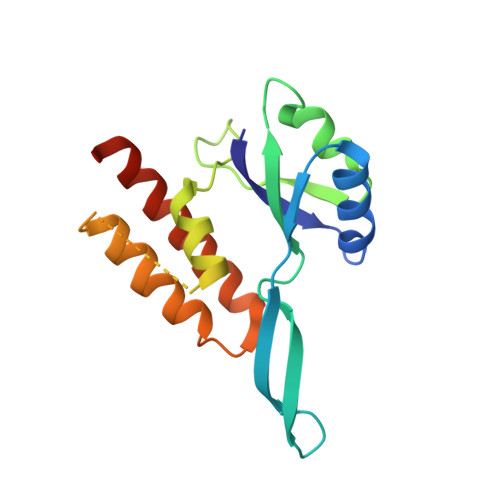
RfaH, a paralog of the general transcription factor NusG, is recruited to elongating RNA polymerase at specific regulatory sites. The X-ray structure of Escherichia coli RfaH reported here reveals two domains. The N-terminal domain displays high similarity to that of NusG. In contrast, the alpha-helical coiled-coil C domain, while retaining sequence similarity, is strikingly different from the beta barrel of NusG. To our knowledge, such an all-beta to all-alpha transition of the entire domain is the most extreme example of protein fold evolution known to date. Both N domains possess a vast hydrophobic cavity that is buried by the C domain in RfaH but is exposed in NusG. We propose that this cavity constitutes the RNA polymerase-binding site, which becomes unmasked in RfaH only upon sequence-specific binding to the nontemplate DNA strand that triggers domain dissociation. Finally, we argue that RfaH binds to the beta' subunit coiled coil, the major target site for the initiation sigma factors.
- Department of Microbiology, The Ohio State University, Columbus, OH 43210, USA.
Organizational Affiliation: