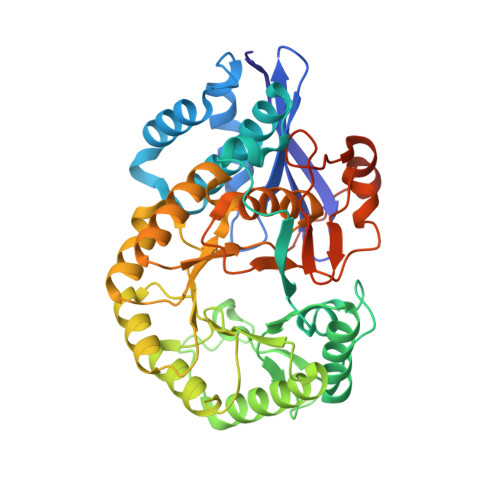
Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400
Bonanno, J.B., Dickey, M., Bain, K.T., Wu, B., Sridhar, V., Freeman, J., Smyth, L., Atwell, S., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.