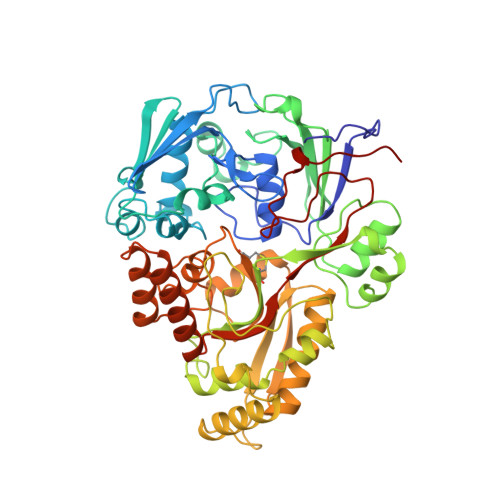
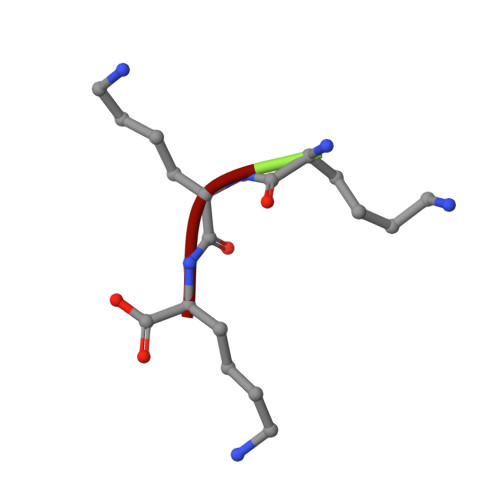
The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands.
Tame, J.R., Dodson, E.J., Murshudov, G., Higgins, C.F., Wilkinson, A.J.(1995) Structure 3: 1395-1406
- PubMed: 8747465
- DOI: https://doi.org/10.1016/s0969-2126(01)00276-3
- Primary Citation of Related Structures:
1OLC, 2OLB - PubMed Abstract:
The periplasmic oligopeptide-binding protein OppA has a remarkably broad substrate specificity, binding peptides of two or five amino-acid residues with high affinity, but little regard to sequence. It is therefore an ideal system for studying how different chemical groups can be accommodated in a protein interior. The ability of the protein to bind peptides of different lengths has been studied by co-crystallising it with different ligands. Crystals of OppA from Salmonella typhimurium complexed with the peptides Lys-Lys-Lys (KKK) and Lys-Lys-Lys-Ala (KKKA) have been grown in the presence of uranyl ions which form important crystal contacts. These structures have been refined to 1.4 A and 2.1 A, respectively. The ligands are completely enclosed, their side chains pointing into large hydrated cavities and making few strong interactions with the protein. Tight peptide binding by OppA arises from strong hydrogen bonding and electrostatic interactions between the protein and the main chain of the ligand. Different basic side chains on the protein form salt bridges with the C terminus of peptide ligands of different lengths.
- Department of Chemistry, University of York, UK.
Organizational Affiliation: