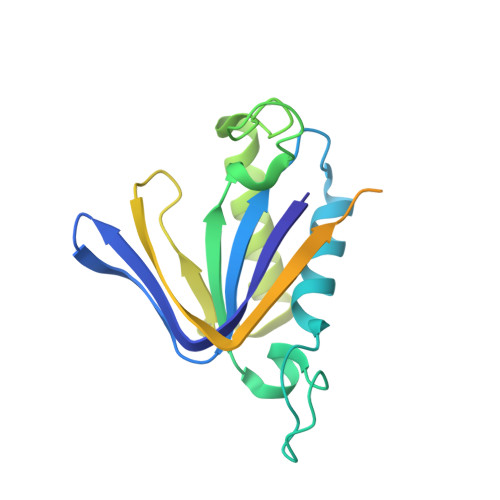
Structure and Function of the c-myc DNA-unwinding Element-binding Protein DUE-B.
Kemp, M., Bae, B., Yu, J.P., Ghosh, M., Leffak, M., Nair, S.K.(2007) J Biological Chem 282: 10441-10448
- PubMed: 17264083
- DOI: https://doi.org/10.1074/jbc.M609632200
- Primary Citation of Related Structures:
2OKV - PubMed Abstract:
Local zones of easily unwound DNA are characteristic of prokaryotic and eukaryotic replication origins. The DNA-unwinding element of the human c-myc replication origin is essential for replicator activity and is a target of the DNA-unwinding element-binding protein DUE-B in vivo. We present here the 2.0A crystal structure of DUE-B and complementary biochemical characterization of its biological activity. The structure corresponds to a dimer of the N-terminal domain of the full-length protein and contains many of the structural elements of the nucleotide binding fold. A single magnesium ion resides in the putative active site cavity, which could serve to facilitate ATP hydrolytic activity of this protein. The structure also demonstrates a notable similarity to those of tRNA-editing enzymes. Consistent with this structural homology, the N-terminal core of DUE-B is shown to display both D-aminoacyl-tRNA deacylase activity and ATPase activity. We further demonstrate that the C-terminal portion of the enzyme is disordered and not essential for dimerization. However, this region is essential for DNA binding in vitro and becomes ordered in the presence of DNA.
- Department of Biochemistry and Molecular Biology, Boonshoft School of Medicine, Wright State University, Dayton, Ohio 45435, USA.
Organizational Affiliation: