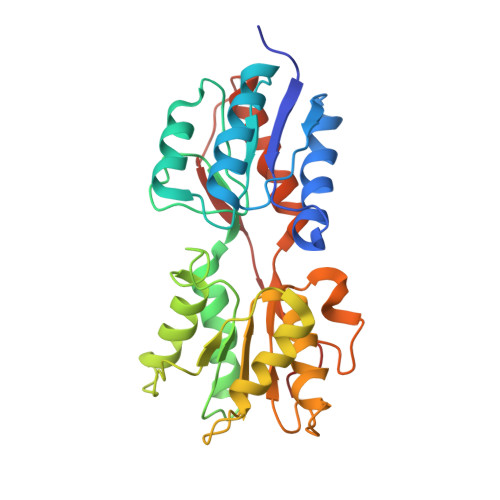
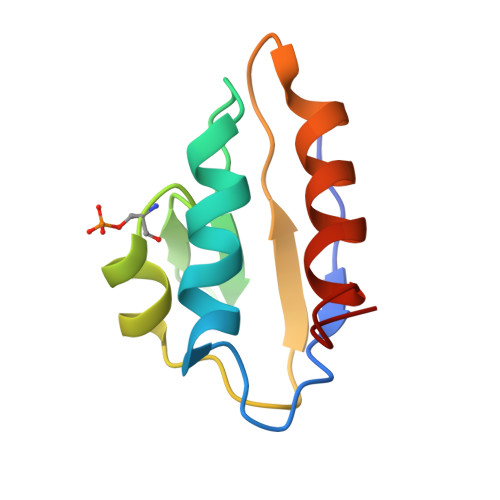
Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
Schumacher, M.A., Seidel, G., Hillen, W., Brennan, R.G.(2007) J Mol Biology 368: 1042-1050
- PubMed: 17376479
- DOI: https://doi.org/10.1016/j.jmb.2007.02.054
- Primary Citation of Related Structures:
2NZU, 2NZV, 2OEN - PubMed Abstract:
In Gram-positive bacteria, carbon catabolite regulation (CCR) is mediated by the carbon catabolite control protein A (CcpA), a member of the LacI-GalR family of transcription regulators. Unlike other LacI-GalR proteins, CcpA is activated to bind DNA by binding the phosphoproteins HPr-Ser46-P or Crh-Ser46-P. However, fine regulation of CCR is accomplished by the small molecule effectors, glucose 6-phosphate (G6P) and fructose 1,6-bisphosphate (FBP), which somehow enhance CcpA-(HPr-Ser46-P) binding to DNA. Unlike the CcpA-(HPr-Ser46-P) complex, DNA binding by CcpA-(Crh-Ser46-P) is not stimulated by G6P or FBP. To understand the fine-tuning mechanism of these effectors, we solved the structures of the CcpA core, DeltaCcpA, which lacks the N-terminal DNA-binding domain, in complex with HPr-Ser46-P and G6P or FBP. G6P and FBP bind in a deep cleft, between the N and C subdomains of CcpA. Neither interacts with HPr-Ser46-P. This suggests that one role of the adjunct corepressors is to buttress the DNA-binding conformation effected by the binding of HPr-Ser46-P to the CcpA dimer N subdomains. However, the structures reveal that an unexpected function of adjunct corepressor binding is to bolster cross interactions between HPr-Ser46-P residue Arg17 and residues Asp69 and Asp99 of the other CcpA subunit. These cross contacts, which are weak or not present in the CcpA-(Crh-Ser46-P) complex, stimulate the CcpA-(HPr-Ser46-P)-DNA interaction specifically. Thus, stabilization of the closed conformation and bolstering of cross contacts between CcpA and its other corepressor, HPr-Ser46-P, provide a molecular explanation for how adjunct corepressors G6P and FBP enhance the interaction between CcpA-(HPr-Ser46-P) and cognate DNA.
- Department of Biochemistry and Molecular Biology, Unit 1000, University of Texas, MD Anderson Cancer Center University, 1515 Holcombe Boulevard, Houston, TX 77030, USA. maschuma@mdanderson.org
Organizational Affiliation: