Isolation, cloning and structural characterisation of boophilin, a multifunctional kunitz-type proteinase inhibitor from the cattle tick.
Macedo-Ribeiro, S., Almeida, C., Calisto, B.M., Friedrich, T., Mentele, R., Sturzebecher, J., Fuentes-Prior, P., Pereira, P.J.(2008) PLoS One 3: e1624-e1624
- PubMed: 18286181
- DOI: https://doi.org/10.1371/journal.pone.0001624
- Primary Citation of Related Structures:
2ODY - PubMed Abstract:
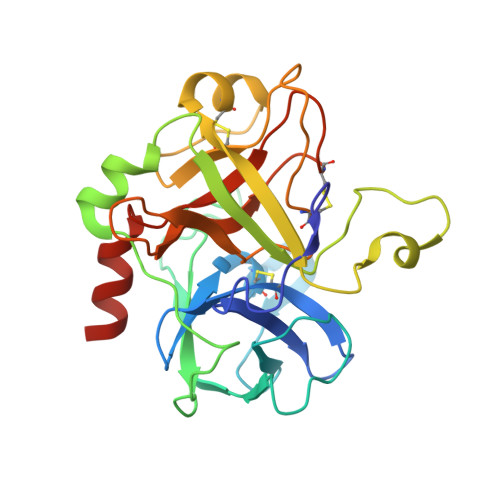
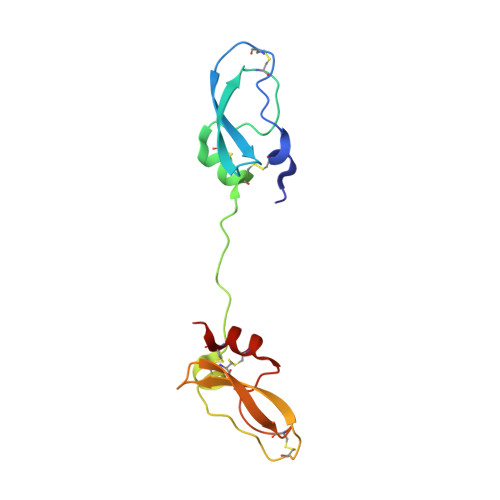
Inhibitors of coagulation factors from blood-feeding animals display a wide variety of structural motifs and inhibition mechanisms. We have isolated a novel inhibitor from the cattle tick Boophilus microplus, one of the most widespread parasites of farm animals. The inhibitor, which we have termed boophilin, has been cloned and overexpressed in Escherichia coli. Mature boophilin is composed of two canonical Kunitz-type domains, and inhibits not only the major procoagulant enzyme, thrombin, but in addition, and by contrast to all other previously characterised natural thrombin inhibitors, significantly interferes with the proteolytic activity of other serine proteinases such as trypsin and plasmin. The crystal structure of the bovine alpha-thrombin.boophilin complex, refined at 2.35 A resolution reveals a non-canonical binding mode to the proteinase. The N-terminal region of the mature inhibitor, Q16-R17-N18, binds in a parallel manner across the active site of the proteinase, with the guanidinium group of R17 anchored in the S(1) pocket, while the C-terminal Kunitz domain is negatively charged and docks into the basic exosite I of thrombin. This binding mode resembles the previously characterised thrombin inhibitor, ornithodorin which, unlike boophilin, is composed of two distorted Kunitz modules. Unexpectedly, both boophilin domains adopt markedly different orientations when compared to those of ornithodorin, in its complex with thrombin. The N-terminal boophilin domain rotates 9 degrees and is displaced by 6 A, while the C-terminal domain rotates almost 6 degrees accompanied by a 3 A displacement. The reactive-site loop of the N-terminal Kunitz domain of boophilin with its P(1) residue, K31, is fully solvent exposed and could thus bind a second trypsin-like proteinase without sterical restraints. This finding explains the formation of a ternary thrombin.boophilin.trypsin complex, and suggests a mechanism for prothrombinase inhibition in vivo.
- Centro de Neurociências e Biologia Celular (CNC), Coimbra, Portugal.
Organizational Affiliation: