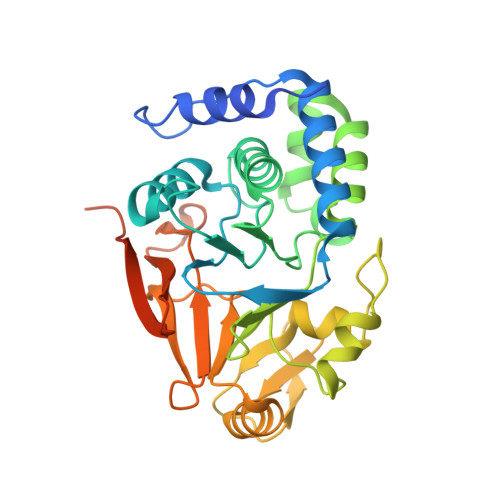
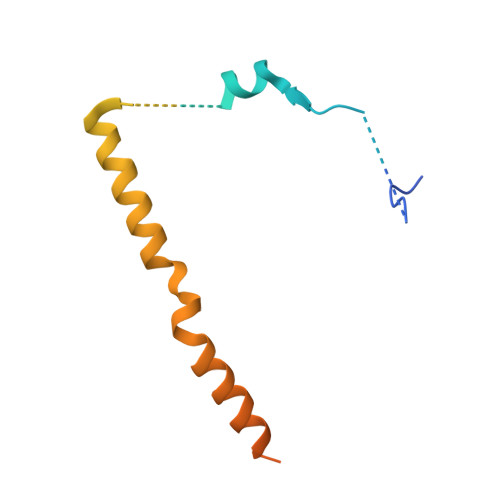
Structural basis for regulation of protein phosphatase 1 by inhibitor-2.
Hurley, T.D., Yang, J., Zhang, L., Goodwin, K.D., Zou, Q., Cortese, M., Dunker, A.K., Depaoli-Roach, A.A.(2007) J Biological Chem 282: 28874-28883
- PubMed: 17636256
- DOI: https://doi.org/10.1074/jbc.M703472200
- Primary Citation of Related Structures:
2O8A, 2O8G - PubMed Abstract:
The functional specificity of type 1 protein phosphatases (PP1) depends on the associated regulatory/targeting and inhibitory subunits. To gain insights into the mechanism of PP1 regulation by inhibitor-2, an ancient and intrinsically disordered regulator, we solved the crystal structure of the complex to 2.5A resolution. Our studies show that, when complexed with PP1c, I-2 acquires three regions of order: site 1, residues 12-17, binds adjacent to a region recognized by many PP1 regulators; site 2, amino acids 44-56, interacts along the RVXF binding groove through an unsuspected sequence, KSQKW; and site 3, residues 130-169, forms alpha-helical regions that lie across the substrate-binding cleft. Specifically, residues 148-151 interact at the catalytic center, displacing essential metal ions, accounting for both rapid inhibition and slower inactivation of PP1c. Thus, our structure provides novel insights into the mechanism of PP1 inhibition and subsequent reactivation, has broad implications for the physiological regulation of PP1, and highlights common inhibitory interactions among phosphoprotein phosphatase family members.
- Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, Indiana 46202. Electronic address: thurley@iupui.edu.
Organizational Affiliation: