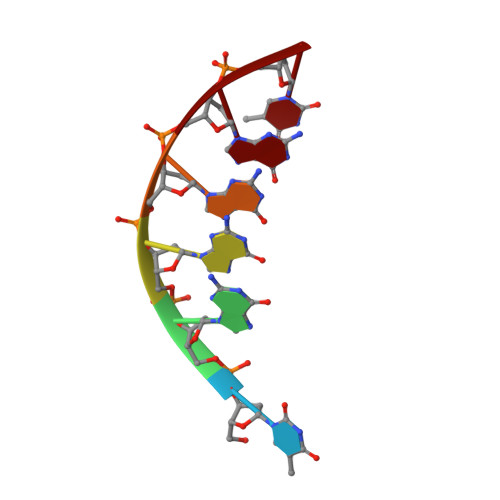
Structure of a d(TGGGGT) quadruplex crystallized in the presence of Li+ ions.
Creze, C., Rinaldi, B., Haser, R., Bouvet, P., Gouet, P.(2007) Acta Crystallogr D Biol Crystallogr 63: 682-688
- PubMed: 17505106
- DOI: https://doi.org/10.1107/S0907444907013315
- Primary Citation of Related Structures:
2O4F - PubMed Abstract:
A parallel 5'-d(TGGGGT)-3' quadruplex was formed in Na(+) solution and crystallized using lithium sulfate as the main precipitating agent. The X-ray structure was determined to 1.5 A resolution in space group P2(1) by molecular replacement. The asymmetric unit consists of a characteristic motif of two quadruplexes stacked at their 5' ends. All nucleotides are clearly defined in the density and could be positioned. A single bound Li(+) ion is observed at the surface of the column formed by the two joined molecules. Thus, this small alkali metal ion appears to be unsuitable as a replacement for the Na(+) ion in the central channel of G-quartets, unlike K(+) or Tl(+) ions. A well conserved constellation of water molecules is observed in the grooves of the dimeric structure.
- Laboratoire de BioCristallographie, Institut de Biologie et Chimie des Protéines, CNRS-UCBL, UMR 5086, IFR128 BioSciences Lyon-Gerland, 7 Passage du Vercors, 69007 Lyon, France.
Organizational Affiliation: