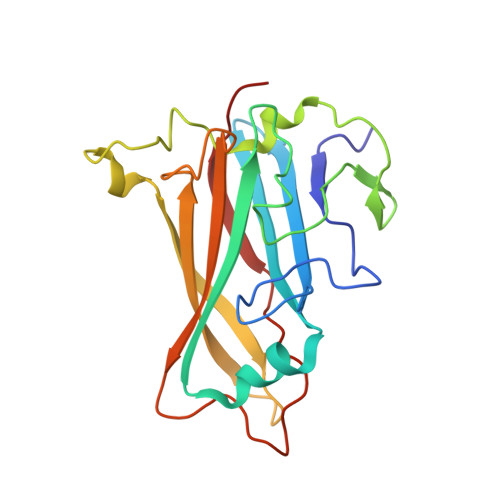
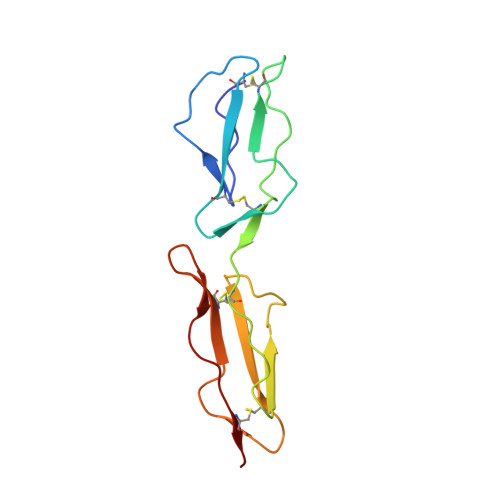
Adenovirus type 11 binding alters the conformation of its receptor CD46.
Persson, B.D., Reiter, D.M., Marttila, M., Mei, Y.F., Casasnovas, J.M., Arnberg, N., Stehle, T.(2007) Nat Struct Mol Biol 14: 164-166
- PubMed: 17220899
- DOI: https://doi.org/10.1038/nsmb1190
- Primary Citation of Related Structures:
2O39 - PubMed Abstract:
Adenoviruses (Ads) are important human pathogens and valuable gene delivery vehicles. We report here the crystal structure of the species B Ad11 knob complexed with the Ad11-binding region of its receptor CD46. The conformation of bound CD46 differs profoundly from its unbound state, with the bent surface structure straightened into an elongated rod. This mechanism of interaction is likely to be conserved among many pathogens that target CD46 or related molecules.
- Interfaculty Institute for Biochemistry, University of Tübingen, D-72076 Tübingen, Germany.
Organizational Affiliation: