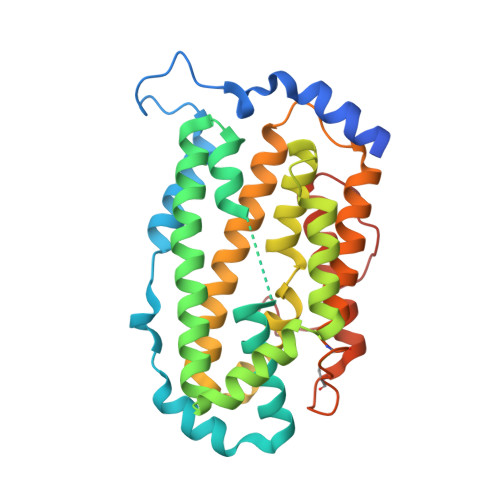
Crystal structure of Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155)
DONG, A., TEMPEL, W., QIU, W., LEW, J., WERNIMONT, A.K., LIN, Y.H., HASSANALI, A., MELONE, M., ZHAO, Y., NORDLUND, P., Arrowsmith, C.H., Edwards, A.M., Weigelt, J., Sundstrom, M., Bochkarev, A., Hui, R., Artz, J.D., Amani, M.To be published.