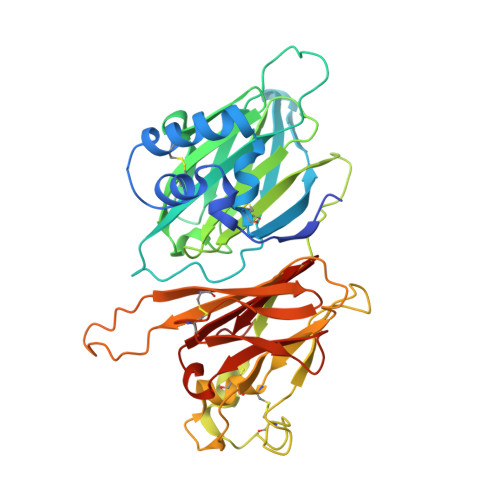
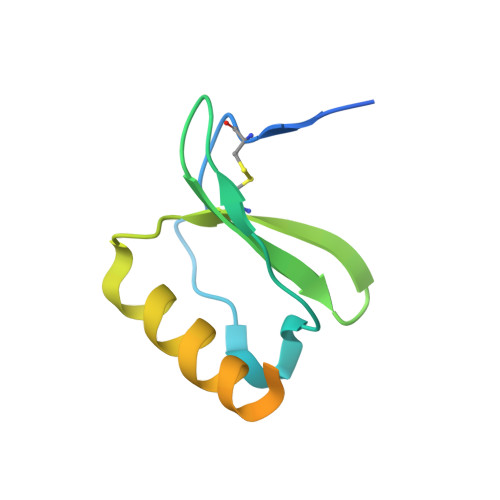
Dual GPCR and GAG mimicry by the M3 chemokine decoy receptor.
Alexander-Brett, J.M., Fremont, D.H.(2007) J Exp Medicine 204: 3157-3172
- PubMed: 18070938
- DOI: https://doi.org/10.1084/jem.20071677
- Primary Citation of Related Structures:
2NYZ, 2NZ1 - PubMed Abstract:
Viruses have evolved a myriad of evasion strategies focused on undermining chemokine-mediated immune surveillance, exemplified by the mouse gamma-herpesvirus 68 M3 decoy receptor. Crystal structures of M3 in complex with C chemokine ligand 1/lymphotactin and CC chemokine ligand 2/monocyte chemoattractant protein 1 reveal that invariant chemokine features associated with G protein-coupled receptor binding are primarily recognized by the decoy C-terminal domain, whereas the N-terminal domain (NTD) reconfigures to engage divergent basic residue clusters on the surface of chemokines. Favorable electrostatic forces dramatically enhance the association kinetics of chemokine binding by M3, with a primary role ascribed to acidic NTD regions that effectively mimic glycosaminoglycan interactions. Thus, M3 employs two distinct mechanisms of chemical imitation to potently sequester chemokines, thereby inhibiting chemokine receptor binding events as well as the formation of chemotactic gradients necessary for directed leukocyte trafficking.
- Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO 63110, USA.
Organizational Affiliation: