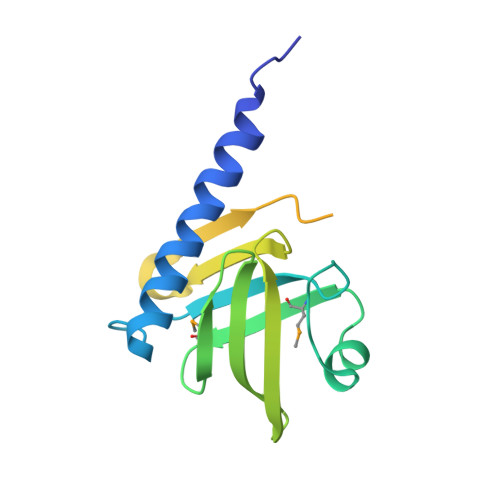
Crystal structure of the hypothetical protein AGR_C_3712 from Agrobacterium tumefaciens.
Vorobiev, S.M., Abashidze, M., Seetharaman, J., Zhao, L., Ma, L.C., Cunningham, K., Nwosu, C., Acton, T.B., Xiao, R., Montelione, G.T., Tong, L., Hunt, J.F.To be published.