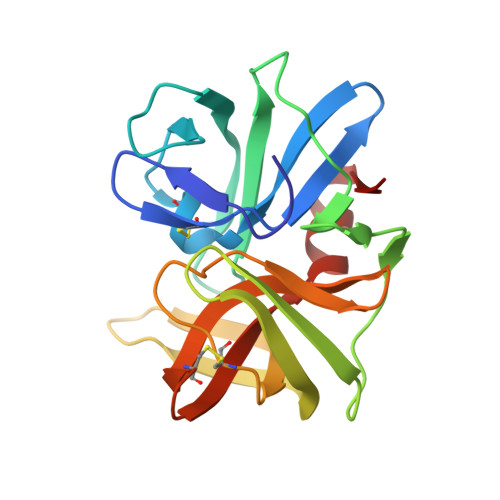
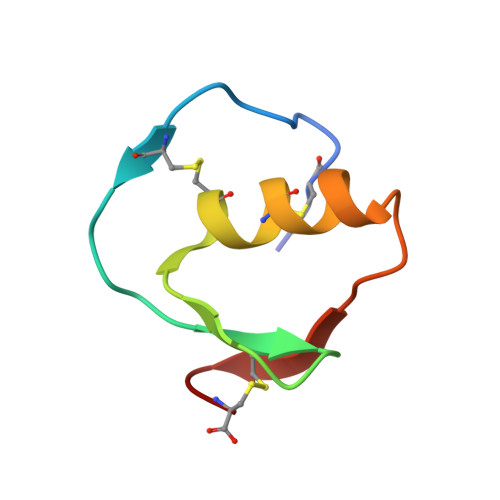
Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
Bateman, K.S., Anderson, S., Lu, W., Qasim, M.A., Laskowski Jr., M., James, M.N.G.To be published.