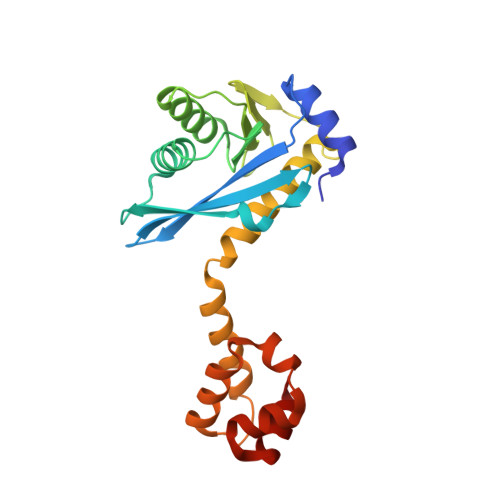
Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad.
Karakas, E., Truglio, J.J., Croteau, D., Rhau, B., Wang, L., Van Houten, B., Kisker, C.(2007) EMBO J 26: 613-622
- PubMed: 17245438
- DOI: https://doi.org/10.1038/sj.emboj.7601497
- Primary Citation of Related Structures:
2NRR, 2NRT, 2NRV, 2NRW, 2NRX, 2NRZ - PubMed Abstract:
Removal and repair of DNA damage by the nucleotide excision repair pathway requires two sequential incision reactions, which are achieved by the endonuclease UvrC in eubacteria. Here, we describe the crystal structure of the C-terminal half of UvrC, which contains the catalytic domain responsible for 5' incision and a helix-hairpin-helix-domain that is implicated in DNA binding. Surprisingly, the 5' catalytic domain shares structural homology with RNase H despite the lack of sequence homology and contains an uncommon DDH triad. The structure also reveals two highly conserved patches on the surface of the protein, which are not related to the active site. Mutations of residues in one of these patches led to the inability of the enzyme to bind DNA and severely compromised both incision reactions. Based on our results, we suggest a model of how UvrC forms a productive protein-DNA complex to excise the damage from DNA.
- Department of Pharmacological Sciences, State University of New York at Stony Brook, Stony Brook, NY, USA.
Organizational Affiliation: