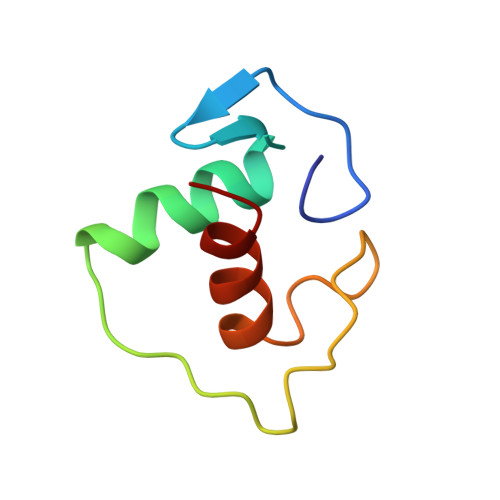
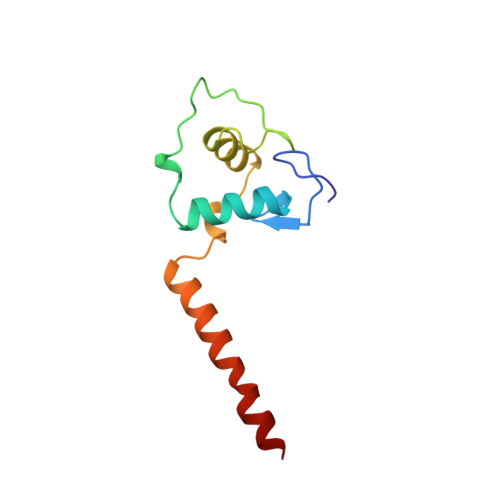
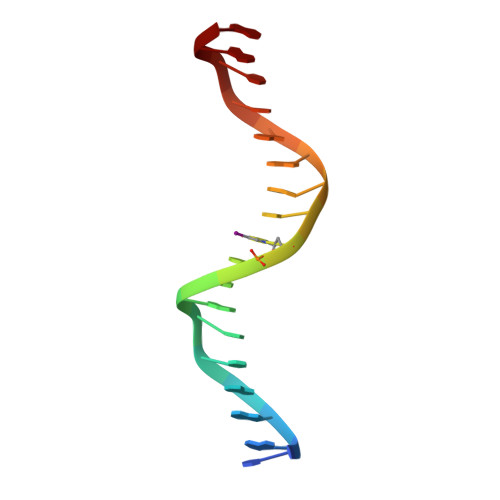
Structural determinants of nuclear receptor assembly on DNA direct repeats.
Rastinejad, F., Perlmann, T., Evans, R.M., Sigler, P.B.(1995) Nature 375: 203-211
- PubMed: 7746322
- DOI: https://doi.org/10.1038/375203a0
- Primary Citation of Related Structures:
2NLL - PubMed Abstract:
Nuclear receptor heterodimers recognize response elements composed of two direct repeats of the consensus sequence 5'-AGGTCA-3' separated by one to five base pairs. The 1.9 A crystal structure of the complex formed by the DNA-binding domains of the 9-cis retinoic acid receptor and thyroid hormone receptor bound to a thyroid-response element shows that the subunits interact through a DNA-supported interface involving the carboxy-terminal extension of the DNA-binding domain of the thyroid hormone receptor. The stereochemistry suggests a mechanism by which heterodimers recognize the inter-half-site spacing between direct repeats.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06510, USA.
Organizational Affiliation: