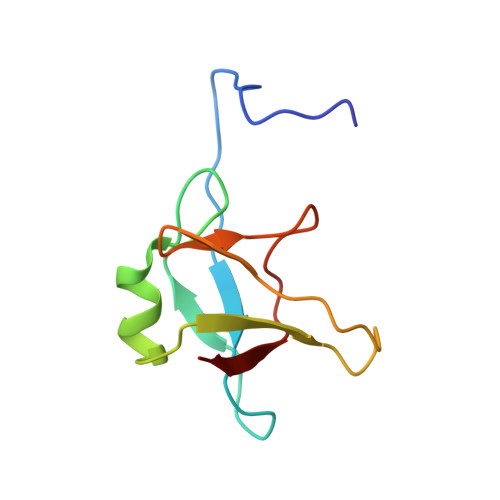
The TDP-43 N-terminal domain structure at high resolution.
Mompean, M., Romano, V., Pantoja-Uceda, D., Stuani, C., Baralle, F.E., Buratti, E., Laurents, D.V.(2016) FEBS J 283: 1242-1260
- PubMed: 26756435
- DOI: https://doi.org/10.1111/febs.13651
- Primary Citation of Related Structures:
2N4P - PubMed Abstract:
Transactive response DNA-binding protein 43 kDa (TDP-43) is an RNA transporting and processing protein whose aberrant aggregates are implicated in neurodegenerative diseases. The C-terminal domain of this protein plays a key role in mediating this process. However, the N-terminal domain (residues 1-77) is needed to effectively recruit TDP-43 monomers into this aggregate. In the present study, we report, for the first time, the essentially complete (1) H, (15) N and (13) C NMR assignments and the structure of the N-terminal domain determined on the basis of 26 hydrogen-bond, 60 torsion angle and 1058 unambiguous NOE structural restraints. The structure consists of an α-helix and six β-strands. Two β-strands form a β-hairpin not seen in the ubiquitin fold. All Pro residues are in the trans conformer and the two Cys are reduced and distantly separated on the surface of the protein. The domain has a well defined hydrophobic core composed of F35, Y43, W68, Y73 and 17 aliphatic side chains. The fold is topologically similar to the reported structure of axin 1. The protein is stable and no denatured species are observed at pH 4 and 25 °C. At 4 kcal·mol(-1) , the conformational stability of the domain, as measured by hydrogen/deuterium exchange, is comparable to ubiquitin (6 kcal·mol(-1) ). The β-strands, α-helix, and three of four turns are generally rigid, although the loop formed by residues 47-53 is mobile, as determined by model-free analysis of the (15) N{(1) H}NOE, as well as the translational and transversal relaxation rates. Structural data have been deposited in the Protein Data Bank under accession code: 2n4p. The NMR assignments have been deposited in the BMRB database under access code: 25675.
- Instituto de Química Física Rocasolano, Madrid, Spain.
Organizational Affiliation: