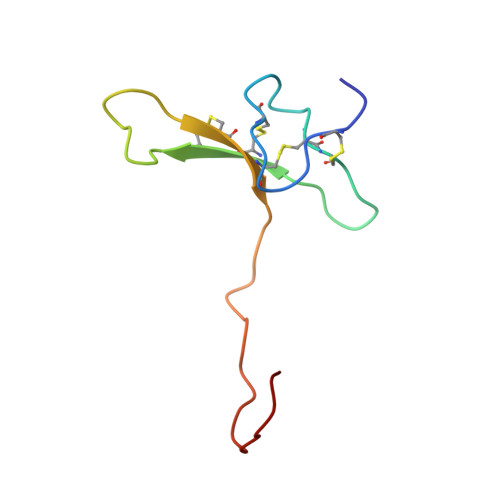
Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Oparin, P.B., Nadezhdin, K.D., Berkut, A.A., Arseniev, A.S., Grishin, E.V., Vassilevski, A.A.(2016) Biochem J 473: 3113-3126
- PubMed: 27412961
- DOI: https://doi.org/10.1042/BCJ20160573
- Primary Citation of Related Structures:
2MZF, 2MZG - PubMed Abstract:
Traditionally, arachnid venoms are known to contain two particularly important groups of peptide toxins. One is disulfide-rich neurotoxins with a predominance of β-structure that specifically target protein receptors in neurons or muscle cells. The other is linear cationic cytotoxins that form amphiphilic α-helices and exhibit rather non-specific membrane-damaging activity. In the present paper, we describe the first 3D structure of a modular arachnid toxin, purotoxin-2 (PT2) from the wolf spider Alopecosa marikovskyi (Lycosidae), studied by NMR spectroscopy. PT2 is composed of an N-terminal inhibitor cystine knot (ICK, or knottin) β-structural domain and a C-terminal linear cationic domain. In aqueous solution, the C-terminal fragment is hyper-flexible, whereas the knottin domain is very rigid. In membrane-mimicking environment, the C-terminal domain assumes a stable amphipathic α-helix. This helix effectively tethers the toxin to membranes and serves as a membrane-access and membrane-anchoring device. Sequence analysis reveals that the knottin + α-helix architecture is quite widespread among arachnid toxins, and PT2 is therefore the founding member of a large family of polypeptides with similar structure motifs. Toxins from this family target different membrane receptors such as P2X in the case of PT2 and calcium channels, but their mechanism of action through membrane access may be strikingly similar.
- M.M. Shemyakin and Yu.A. Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 16/10 Miklukho-Maklaya, Moscow 117997, Russia.
Organizational Affiliation: