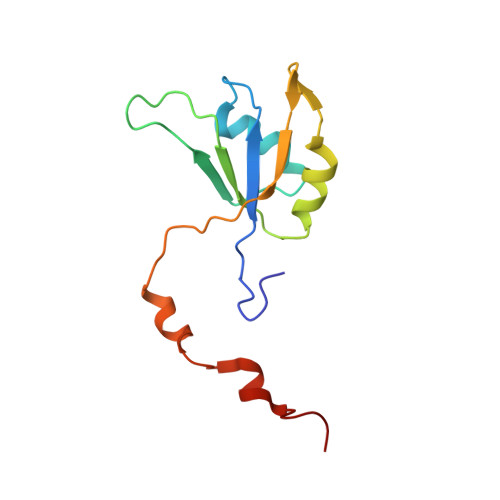
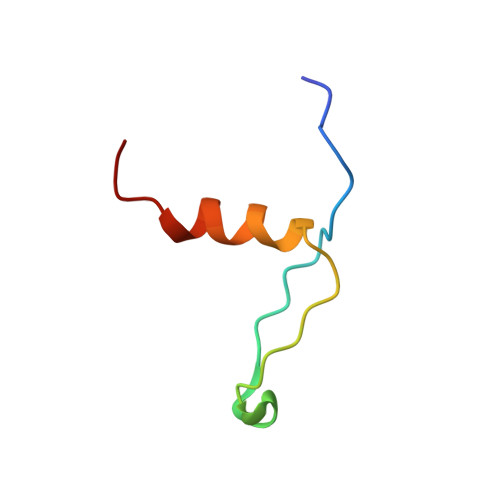
Structures of intermediates during RES complex assembly.
Wysoczanski, P., Becker, S., Zweckstetter, M.(2015) Sci Rep 5: 12545-12545
- PubMed: 26212312
- DOI: https://doi.org/10.1038/srep12545
- Primary Citation of Related Structures:
2MY2, 2MY3 - PubMed Abstract:
The action of the spliceosome depends on the stepwise cooperative assembly and disassembly of its components. Very strong cooperativity was observed for the RES (Retention and Splicing) hetero-trimeric complex where the affinity from binary to tertiary interactions changes more than 100-fold and affects RNA binding. The RES complex is involved in splicing regulation and retention of not properly spliced pre-mRNA with its three components--Snu17p, Pml1p and Bud13p--giving rise to the two possible intermediate dimeric complexes Pml1p-Snu17p and Bud13p-Snu17p. Here we determined the three-dimensional structure and dynamics of the Pml1p-Snu17p and Bud13p-Snu17p dimers using liquid state NMR. We demonstrate that localized as well as global changes occur along the RES trimer assembly pathway. The stepwise rigidification of the Snu17p structure following the binding of Pml1p and Bud13p provides a basis for the strong cooperative nature of RES complex assembly.
- Department for NMR-based Structural Biology, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.
Organizational Affiliation: