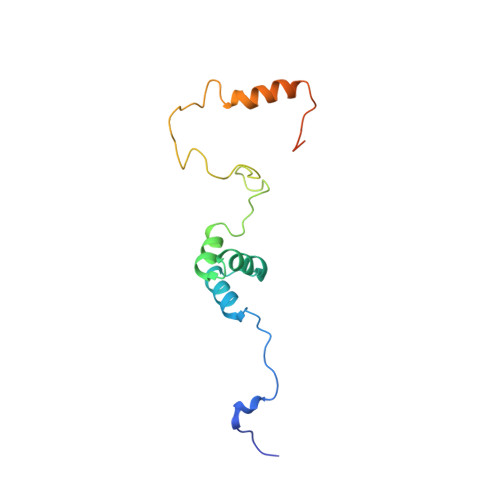
The N-terminal ubiquitin-binding region of ubiquitin-specific protease 28 modulates its deubiquitination function: NMR structural and mechanistic insights.
Wen, Y., Shi, L., Ding, Y., Cui, R., He, W.T., Hu, H.Y., Zhang, N.(2015) Biochem J 471: 155-165
- PubMed: 26268556
- DOI: https://doi.org/10.1042/BJ20150088
- Primary Citation of Related Structures:
2MUU - PubMed Abstract:
The deubiquitinase ubiquitin-specific protease 28 (Usp28) contains a ubiquitin-binding region (UBR) composed of one ubiquitin-associated domain (UBA) and one ubiquitin-interacting motif (UIM) at its N-terminus. It is of interest that an additional small ubiquitin-like modifier (SUMO)-interacting motif (SIM) is located next to its UIM. To date, the functional role of the Usp28 UBR is still not understood. To elucidate the regulatory mechanism of the UBR on the full functional display of Usp28, in the present study, NMR and biochemical approaches were applied. The solution structure of Usp28 UBR was obtained, and the key residues responsible for ubiquitin and SUMO1/2 recognition were identified. In addition, we find that the ubiquitin-binding ability of Usp28 UBR was required for full enzymatic activity of Usp28, whereas binding of SUMO1/2 impaired the catalytic activity of the enzyme by competitively blocking its interactions with ubiquitin substrates. Our findings provide a first insight into understanding how the enzymatic activity of Usp28 is regulated by its non-catalytic UBR and endogenous ligands.
- Department of Analytical Chemistry, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: