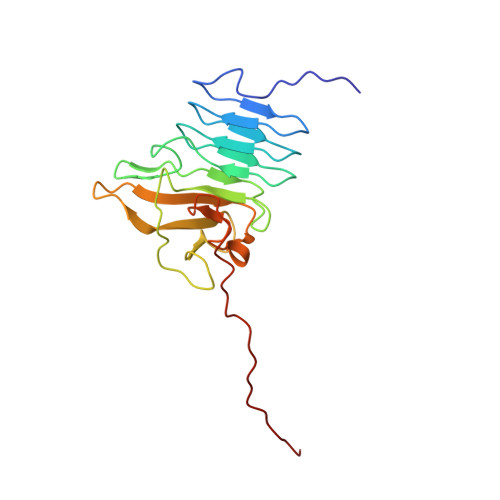
Structural and Functional Characterization of the R-modules in Alginate C-5 Epimerases AlgE4 and AlgE6 from Azotobacter vinelandii
Buchinger, E., Knudsen, D.H., Behrens, M.A., Pedersen, J.S., Aarstad, O.A., Tndervik, A., Valla, S., Skjak-Brk, G., Wimmer, R., Aachmann, F.L.(2014) J Biological Chem 289: 31382-31396
- PubMed: 25266718
- DOI: https://doi.org/10.1074/jbc.M114.567008
- Primary Citation of Related Structures:
2ML1, 2ML2, 2ML3 - PubMed Abstract:
The bacterium Azotobacter vinelandii produces a family of seven secreted and calcium-dependent mannuronan C-5 epimerases (AlgE1-7). These epimerases are responsible for the epimerization of β-D-mannuronic acid (M) to α-L-guluronic acid (G) in alginate polymers. The epimerases display a modular structure composed of one or two catalytic A-modules and from one to seven R-modules having an activating effect on the A-module. In this study, we have determined the NMR structure of the three individual R-modules from AlgE6 (AR1R2R3) and the overall structure of both AlgE4 (AR) and AlgE6 using small angle x-ray scattering. Furthermore, the alginate binding ability of the R-modules of AlgE4 and AlgE6 has been studied with NMR and isothermal titration calorimetry. The AlgE6 R-modules fold into an elongated parallel β-roll with a shallow, positively charged groove across the module. Small angle x-ray scattering analyses of AlgE4 and AlgE6 show an overall elongated shape with some degree of flexibility between the modules for both enzymes. Titration of the R-modules with defined alginate oligomers shows strong interaction between AlgE4R and both oligo-M and MG, whereas no interaction was detected between these oligomers and the individual R-modules from AlgE6. A combination of all three R-modules from AlgE6 shows weak interaction with long M-oligomers. Exchanging the R-modules between AlgE4 and AlgE6 resulted in a novel epimerase called AlgE64 with increased G-block forming ability compared with AlgE6.
- From the Department of Biotechnology, Chemistry and Environmental Engineering, Aalborg University, Frederik Bajers vej 7H, DK-9220 Aalborg, Denmark, the Department of Biotechnology, Norwegian Biopolymer Laboratory (NOBIPOL), Norwegian University of Science and Technology, Sem Sælands vei 6/8, 7491 Trondheim, Norway.
Organizational Affiliation: