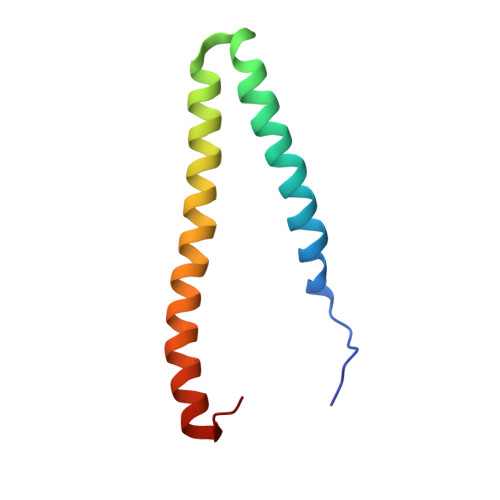
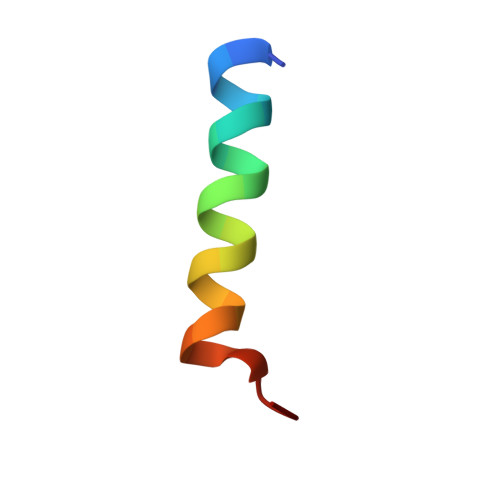
STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry.
Stathopulos, P.B., Schindl, R., Fahrner, M., Zheng, L., Gasmi-Seabrook, G.M., Muik, M., Romanin, C., Ikura, M.(2013) Nat Commun 4: 2963-2963
- PubMed: 24351972
- DOI: https://doi.org/10.1038/ncomms3963
- Primary Citation of Related Structures:
2MAJ, 2MAK - PubMed Abstract:
Orai1 calcium channels in the plasma membrane are activated by stromal interaction molecule-1 (STIM1), an endoplasmic reticulum calcium sensor, to mediate store-operated calcium entry (SOCE). The cytosolic region of STIM1 contains a long putative coiled-coil (CC)1 segment and shorter CC2 and CC3 domains. Here we present solution nuclear magnetic resonance structures of a trypsin-resistant CC1-CC2 fragment in the apo and Orai1-bound states. Each CC1-CC2 subunit forms a U-shaped structure that homodimerizes through antiparallel interactions between equivalent α-helices. The CC2:CC2' helix pair clamps two identical acidic Orai1 C-terminal helices at opposite ends of a hydrophobic/basic STIM-Orai association pocket. STIM1 mutants disrupting CC1:CC1' interactions attenuate, while variants promoting CC1 stability spontaneously activate Orai1 currents. CC2 mutations cause remarkable variability in Orai1 activation because of a dual function in binding Orai1 and autoinhibiting STIM1 oligomerization via interactions with CC3. We conclude that SOCE is activated through dynamic interplay between STIM1 and Orai1 helices.
- University Health Network and Department of Medical Biophysics, Campbell Family Cancer Research Institute, Ontario Cancer Institute, University of Toronto, Room 4-804, MaRS TMDT, 101 College Street, Toronto, Ontario, Canada M5G 1L7.
Organizational Affiliation: