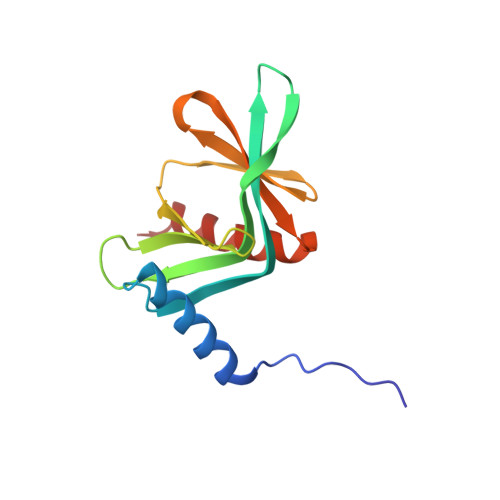
A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5' exonucleolytic degradation.
Braun, J.E., Truffault, V., Boland, A., Huntzinger, E., Chang, C.T., Haas, G., Weichenrieder, O., Coles, M., Izaurralde, E.(2012) Nat Struct Mol Biol 19: 1324-1331
- PubMed: 23142987
- DOI: https://doi.org/10.1038/nsmb.2413
- Primary Citation of Related Structures:
2LYD - PubMed Abstract:
The removal of the mRNA 5' cap structure by the decapping enzyme DCP2 leads to rapid 5'→3' mRNA degradation by XRN1, suggesting that the two processes are coordinated, but the coupling mechanism is unknown. DCP2 associates with the decapping activators EDC4 and DCP1. Here we show that XRN1 directly interacts with EDC4 and DCP1 in human and Drosophila melanogaster cells, respectively. In D. melanogaster cells, this interaction is mediated by the DCP1 EVH1 domain and a DCP1-binding motif (DBM) in the XRN1 C-terminal region. The NMR structure of the DCP1 EVH1 domain bound to the DBM reveals that the peptide docks at a conserved aromatic cleft, which is used by EVH1 domains to recognize proline-rich ligands. Our findings reveal a role for XRN1 in decapping and provide a molecular basis for the coupling of decapping to 5'→3' mRNA degradation.
- Department of Biochemistry, Max Planck Institute for Developmental Biology, Tübingen, Germany.
Organizational Affiliation: