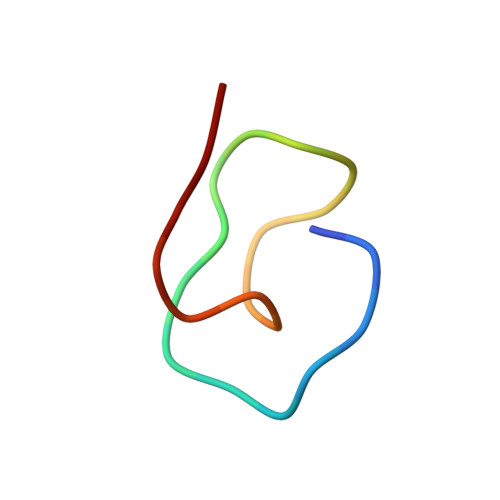
Precursor-centric genome-mining approach for lasso peptide discovery.
Maksimov, M.O., Pelczer, I., Link, A.J.(2012) Proc Natl Acad Sci U S A 109: 15223-15228
- PubMed: 22949633
- DOI: https://doi.org/10.1073/pnas.1208978109
- Primary Citation of Related Structures:
2LTI - PubMed Abstract:
Lasso peptides are a class of ribosomally synthesized posttranslationally modified natural products found in bacteria. Currently known lasso peptides have a diverse set of pharmacologically relevant activities, including inhibition of bacterial growth, receptor antagonism, and enzyme inhibition. The biosynthesis of lasso peptides is specified by a cluster of three genes encoding a precursor protein and two enzymes. Here we develop a unique genome-mining algorithm to identify lasso peptide gene clusters in prokaryotes. Our approach involves pattern matching to a small number of conserved amino acids in precursor proteins, and thus allows for a more global survey of lasso peptide gene clusters than does homology-based genome mining. Of more than 3,000 currently sequenced prokaryotic genomes, we found 76 organisms that are putative lasso peptide producers. These organisms span nine bacterial phyla and an archaeal phylum. To provide validation of the genome-mining method, we focused on a single lasso peptide predicted to be produced by the freshwater bacterium Asticcacaulis excentricus. Heterologous expression of an engineered, minimal gene cluster in Escherichia coli led to the production of a unique lasso peptide, astexin-1. At 23 aa, astexin-1 is the largest lasso peptide isolated to date. It is also highly polar, in contrast to many lasso peptides that are primarily hydrophobic. Astexin-1 has modest antimicrobial activity against its phylogenetic relative Caulobacter crescentus. The solution structure of astexin-1 was determined revealing a unique topology that is stabilized by hydrogen bonding between segments of the peptide.
- Department of Chemical and Biological Engineering, Princeton University, Princeton, NJ 08544, USA.
Organizational Affiliation: