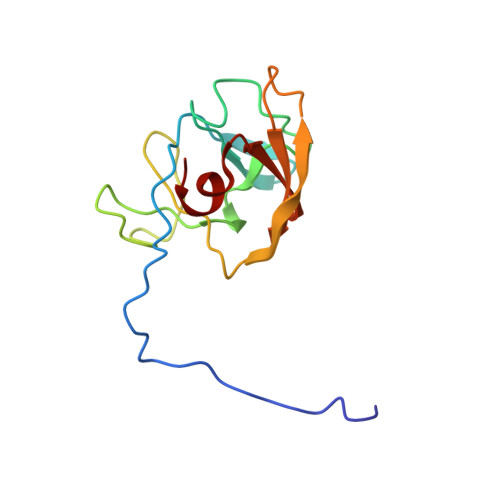
Solution structure of the recombinant target recognition domain of zoocin A.
Chen, Y., Simmonds, R.S., Young, J.K., Timkovich, R.(2013) Proteins 81: 722-727
- PubMed: 23184858
- DOI: https://doi.org/10.1002/prot.24224
- Primary Citation of Related Structures:
2LS0 - PubMed Abstract:
The protein rTRD is the recombinant form of the target recognition domain of zoocin A, a lytic exoenzyme produced by Streptococcus equi subspecies zooepidemicus 4881. It has no known sequence homologs. However, the catalytic domain of zoocin A is homologous to lysostaphin which is another exoenzyme active against a different spectrum of bacteria, including the pathogen Staphylococcus aureus. An ensemble of models for the solution structure of rTRD has been generated by NMR techniques. The minimum energy model from the ensemble was subjected to three-dimensional homology search engines, but no homologs were found, suggesting rTRD may represent a new protein folding family. There is some similarity in the folding of rTRD to the immunoglobin fold of the antigen binding region of mammalian antibodies which could suggest an ancient evolutionary relation.
- Department of Chemistry, University of Alabama, Tuscaloosa, Alabama 35487-0336, USA.
Organizational Affiliation: