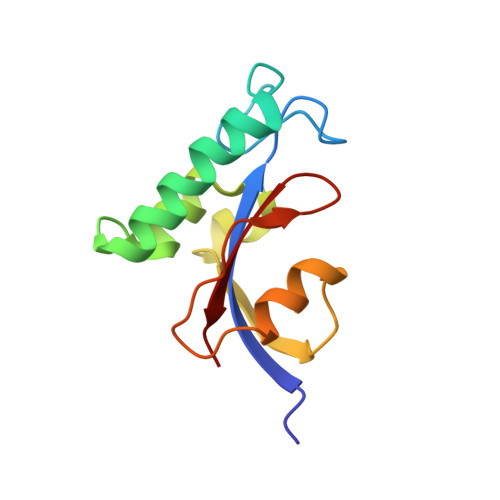
Solution structural analysis of the single-domain parvulin TbPin1.
Sun, L., Wu, X., Peng, Y., Goh, J.Y., Liou, Y.-C., Lin, D., Zhao, Y.(2012) PLoS One 7: e43017-e43017
- PubMed: 22900083
- DOI: https://doi.org/10.1371/journal.pone.0043017
- Primary Citation of Related Structures:
2LJ4 - PubMed Abstract:
Pin1-type parvulins are phosphorylation-dependent peptidyl-prolyl cis-trans isomerases. Their functions have been widely reported to be involved in a variety of cellular responses or processes, such as cell division, transcription, and apoptosis, as well as in human diseases including Alzheimer's disease and cancers. TbPin1 was identified as a novel class of Pin1-type parvulins from Trypanosoma brucei, containing a unique PPIase domain, which can catalyze the isomerization of phosphorylated Ser/Thr-Pro peptide bond. We determined the solution structure of TbPin1 and performed (15)N relaxation measurements to analyze its backbone dynamics using multi-dimensional heteronuclear NMR spectroscopy. The average RMSD values of the 20 lowest energy structures are 0.50±0.05 Å for backbone heavy atoms and 0.85±0.08 Å for all heavy atoms. TbPin1 adopts the typical catalytic tertiary structure of Pin1-type parvulins, which comprises a globular fold with a four-stranded anti-parallel β-sheet core surrounded by three α-helices and one 3(10)-helix. The global structure of TbPin1 is relatively rigid except the active site. The 2D EXSY spectra illustrate that TbPin1 possesses a phosphorylation-dependent PPIase activity. The binding sites of TbPin1 for a phosphorylated peptide substrate {SSYFSG[p]TPLEDDSD} were determined by the chemical shift perturbation approach. Residues Ser15, Arg18, Asn19, Val21, Ser22, Val32, Gly66, Ser67, Met83, Asp105 and Gly107 are involved in substantial contact with the substrate. The solution structure of TbPin1 and the binding sites of the phosphorylated peptide substrate on TbPin1 were determined. The work is helpful for further understanding the molecular basis of the substrate specificity for Pin1-type parvulin family and enzyme catalysis.
- The Key Laboratory of Chemical Biology of Fujian Province, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen, China.
Organizational Affiliation: