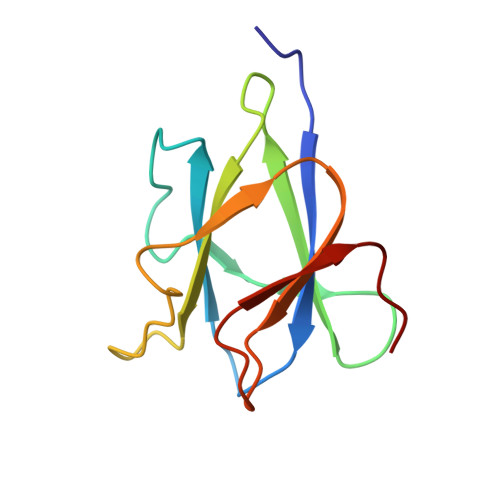
A functional and structural analysis of tundem family 25 carbohydrate-binding modules from Paenibacillus polymyxa beta/alpha-amylase
Takahashi, R., Horibe, I., Fukada, H., Yoshida, T., Ohkubo, T., Inui, T., Nishimura, S., Sumitani, J.To be published.