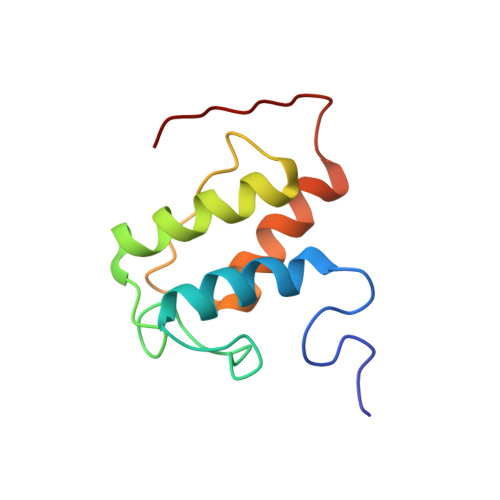
Solution structures of the acyl carrier protein domain from the highly reducing type I iterative polyketide synthase CalE8
Lim, J., Kong, R., Murugan, E., Ho, C.L., Liang, Z.X., Yang, D.(2011) PLoS One 6: e20549-e20549
- PubMed: 21674045
- DOI: https://doi.org/10.1371/journal.pone.0020549
- Primary Citation of Related Structures:
2L9F - PubMed Abstract:
Biosynthesis of the enediyne natural product calicheamicins γ(1) (I) in Micromonospora echinospora ssp. calichensis is initiated by the iterative polyketide synthase (PKS) CalE8. Recent studies showed that CalE8 produces highly conjugated polyenes as potential biosynthetic intermediates and thus belongs to a family of highly-reducing (HR) type I iterative PKSs. We have determined the NMR structure of the ACP domain (meACP) of CalE8, which represents the first structure of a HR type I iterative PKS ACP domain. Featured by a distinct hydrophobic patch and a glutamate-residue rich acidic patch, meACP adopts a twisted three-helix bundle structure rather than the canonical four-helix bundle structure. The so-called 'recognition helix' (α2) of meACP is less negatively charged than the typical type II ACPs. Although loop-2 exhibits greater conformational mobility than other regions of the protein with a missing short helix that can be observed in most ACPs, two bulky non-polar residues (Met(992), Phe(996)) from loop-2 packed against the hydrophobic protein core seem to restrict large movement of the loop and impede the opening of the hydrophobic pocket for sequestering the acyl chains. NMR studies of the hydroxybutyryl- and octanoyl-meACP confirm that meACP is unable to sequester the hydrophobic chains in a well-defined central cavity. Instead, meACP seems to interact with the octanoyl tail through a distinct hydrophobic patch without involving large conformational change of loop-2. NMR titration study of the interaction between meACP and the cognate thioesterase partner CalE7 further suggests that their interaction is likely through the binding of CalE7 to the meACP-tethered polyene moiety rather than direct specific protein-protein interaction.
- Department of Biological Sciences, National University of Singapore, Singapore, Singapore.
Organizational Affiliation: