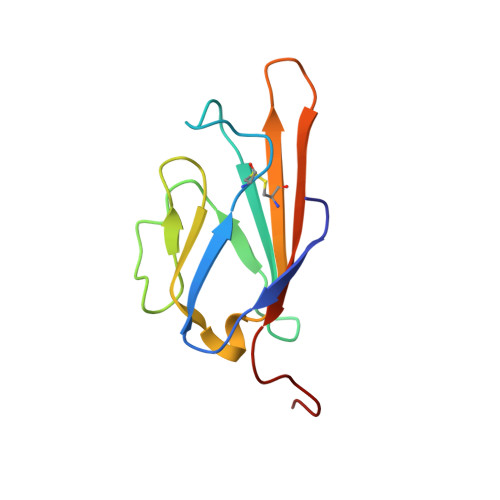
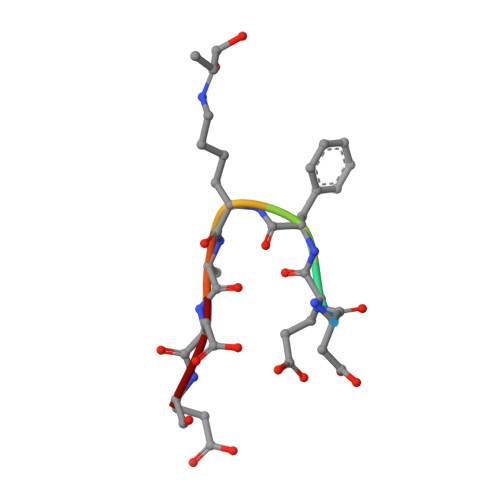
Advanced glycation end product recognition by the receptor for AGEs.
Xue, J., Rai, V., Singer, D., Chabierski, S., Xie, J., Reverdatto, S., Burz, D.S., Schmidt, A.M., Hoffmann, R., Shekhtman, A.(2011) Structure 19: 722-732
- PubMed: 21565706
- DOI: https://doi.org/10.1016/j.str.2011.02.013
- Primary Citation of Related Structures:
2L7U - PubMed Abstract:
Nonenzymatic protein glycation results in the formation of advanced glycation end products (AGEs) that are implicated in the pathology of diabetes, chronic inflammation, Alzheimer's disease, and cancer. AGEs mediate their effects primarily through a receptor-dependent pathway in which AGEs bind to a specific cell surface associated receptor, the Receptor for AGEs (RAGE). N(ɛ)-carboxy-methyl-lysine (CML) and N(ɛ)-carboxy-ethyl-lysine (CEL), constitute two of the major AGE structures found in tissue and blood plasma, and are physiological ligands of RAGE. The solution structure of a CEL-containing peptide-RAGE V domain complex reveals that the carboxyethyl moiety fits inside a positively charged cavity of the V domain. Peptide backbone atoms make specific contacts with the V domain. The geometry of the bound CEL peptide is compatible with many CML (CEL)-modified sites found in plasma proteins. The structure explains how such patterned ligands as CML (CEL)-proteins bind to RAGE and contribute to RAGE signaling.
- Department of Chemistry, State University of New York at Albany, Albany, NY 12222, USA.
Organizational Affiliation: