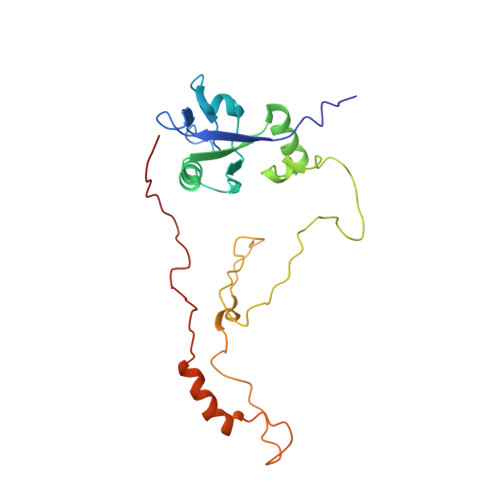
A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation.
Bhattacharya, S., Dai, Z., Li, J., Baxter, S., Callaway, D.J.E., Cowburn, D., Bu, Z.(2010) J Biological Chem 285: 9981-9994
- PubMed: 20042604
- DOI: https://doi.org/10.1074/jbc.M109.074005
- Primary Citation of Related Structures:
2KRG - PubMed Abstract:
The mammalian Na(+)/H(+) exchange regulatory factor 1 (NHERF1) is a multidomain scaffolding protein essential for regulating the intracellular trafficking and macromolecular assembly of transmembrane ion channels and receptors. NHERF1 consists of tandem PDZ-1, PDZ-2 domains that interact with the cytoplasmic domains of membrane proteins and a C-terminal (CT) domain that binds the membrane-cytoskeleton linker protein ezrin. NHERF1 is held in an autoinhibited state through intramolecular interactions between PDZ2 and the CT domain that also includes a C-terminal PDZ-binding motif (-SNL). We have determined the structures of the isolated and tandem PDZ2CT domains by high resolution NMR using small angle x-ray scattering as constraints. The PDZ2CT structure shows weak intramolecular interactions between the largely disordered CT domain and the PDZ ligand binding site. The structure reveals a novel helix-turn-helix subdomain that is allosterically coupled to the putative PDZ2 domain by a network of hydrophobic interactions. This helical subdomain increases both the stability and the binding affinity of the extended PDZ structure. Using NMR and small angle neutron scattering for joint structure refinement, we demonstrate the release of intramolecular domain-domain interactions in PDZ2CT upon binding to ezrin. Based on the structural information, we show that human disease-causing mutations in PDZ2, R153Q and E225K, have significantly reduced protein stability. Loss of NHERF1 expressed in cells could result in failure to assemble membrane complexes that are important for normal physiological functions.
- New York Structural Biology Center, New York, New York 10031.
Organizational Affiliation: