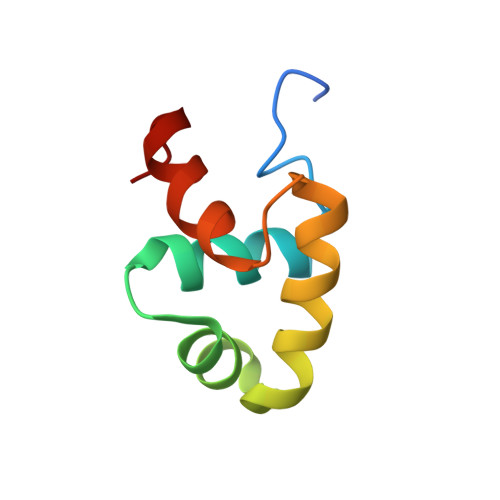
NMR solution structure and DNA-binding model of the DNA-binding domain of competence protein A.
Hobbs, C.A., Bobay, B.G., Thompson, R.J., Perego, M., Cavanagh, J.(2010) J Mol Biology 398: 248-263
- PubMed: 20302877
- DOI: https://doi.org/10.1016/j.jmb.2010.03.003
- Primary Citation of Related Structures:
2KRF - PubMed Abstract:
Competence protein A (ComA) is a response regulator protein involved in the development of genetic competence in the Gram-positive spore-forming bacterium Bacillus subtilis, as well as the regulation of the production of degradative enzymes and antibiotic synthesis. ComA belongs to the NarL family of proteins, which are characterized by a C-terminal transcriptional activator domain that consists of a bundle of four helices, where the second and third helices (alpha 8 and alpha 9) form a helix-turn-helix DNA-binding domain. Using NMR spectroscopy, the high-resolution 3D solution structure of the C-terminal DNA-binding domain of ComA (ComAC) has been determined. In addition, surface plasmon resonance and NMR protein-DNA titration experiments allowed for the analysis of the interaction of ComAC with its target DNA sequences. Combining the solution structure and biochemical data, a model of ComAC bound to the ComA recognition sequences on the srfA promoter has been developed. The model shows that for DNA binding, ComA uses the conserved helix-turn-helix motif present in other NarL family members. However, the model reveals also that ComA might use a slightly different part of the helix-turn-helix motif and there appears to be some associated domain re-orientation. These observations suggest a basis for DNA binding specificity within the NarL family.
- Department of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, NC 27695, USA.
Organizational Affiliation: