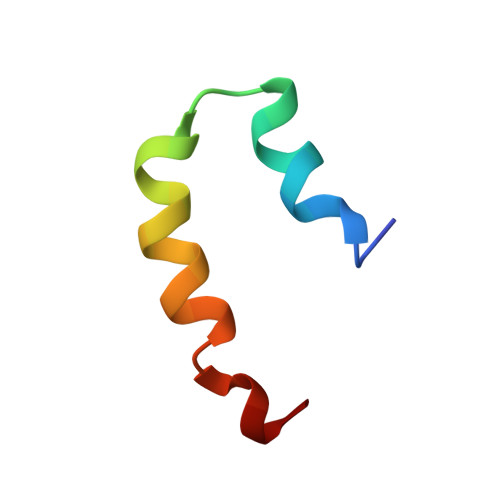
NMR structure of pardaxin, a pore-forming antimicrobial peptide, in lipopolysaccharide Micelles: Mechanism of outer membrane permeabilization
Bhunia, A., Domadia, P.N., Torres, J., Hallock, K.J., Ramamoorthy, A., Bhattacharjya, S.(2010) J Biological Chem 285: 3883-3895
- PubMed: 19959835
- DOI: https://doi.org/10.1074/jbc.M109.065672
- Primary Citation of Related Structures:
2KNS - PubMed Abstract:
Lipopolysaccharide (LPS), the major constituent of the outer membrane of Gram-negative bacteria, is an important element against permeability of bactericidal agents, including antimicrobial peptides. However, structural determinants of antimicrobial peptides for LPS recognition are not clearly understood. Pardaxins (Pa1, Pa2, Pa3, and Pa4) are a group of pore-forming bactericidal peptides found in the mucous glands of sole fishes. Despite having a low net positive charge, pardaxins contain a broad spectrum of antibacterial activities. To elucidate the structural basis of LPS interactions of pardaxins, herein, we report the first three-dimensional structure of Pa4 bound to LPS micelles. The binding kinetics of Pa4 with LPS is estimated using [(15)N-Leu-19] relaxation dispersion NMR experiments. LPS/Pa4 interactions are further characterized by a number of biophysical methods, including isothermal titration calorimetry, (31)P NMR, saturation transfer difference NMR, dynamic light scattering, and IR spectroscopy. In the LPS-Pa4 complex, Pa4 adopts a unique helix-turn-helix conformation resembling a "horseshoe." Interestingly, the LPS-bound structure of Pa4 shows striking differences with the structures determined in lipid micelles or organic solvents. Saturation transfer difference NMR identifies residues of Pa4 that are intimately associated with LPS micelles. Collectively, our results provide mechanistic insights into the outer membrane permeabilization by pardaxin.
- From the School of Biological Sciences, Division of Structural and Computational Biology, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551 and.
Organizational Affiliation: