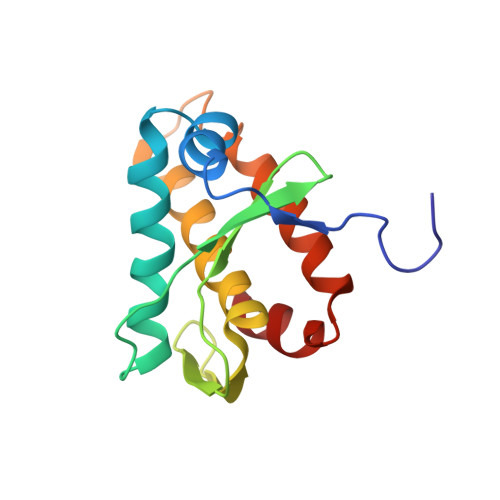
The structure of the KlcA and ArdB proteins reveals a novel fold and antirestriction activity against Type I DNA restriction systems in vivo but not in vitro
Serfiotis-Mitsa, D., Herbert, A.P., Roberts, G.A., Soares, D.C., White, J.H., Blakely, G.W., Uhrin, D., Dryden, D.T.F.(2010) Nucleic Acids Res 38: 1723-1737
- PubMed: 20007596
- DOI: https://doi.org/10.1093/nar/gkp1144
- Primary Citation of Related Structures:
2KMG - PubMed Abstract:
Plasmids, conjugative transposons and phage frequently encode anti-restriction proteins to enhance their chances of entering a new bacterial host that is highly likely to contain a Type I DNA restriction and modification (RM) system. The RM system usually destroys the invading DNA. Some of the anti-restriction proteins are DNA mimics and bind to the RM enzyme to prevent it binding to DNA. In this article, we characterize ArdB anti-restriction proteins and their close homologues, the KlcA proteins from a range of mobile genetic elements; including an ArdB encoded on a pathogenicity island from uropathogenic Escherichia coli and a KlcA from an IncP-1b plasmid, pBP136 isolated from Bordetella pertussis. We show that all the ArdB and KlcA act as anti-restriction proteins and inhibit the four main families of Type I RM systems in vivo, but fail to block the restriction endonuclease activity of the archetypal Type I RM enzyme, EcoKI, in vitro indicating that the action of ArdB is indirect and very different from that of the DNA mimics. We also present the structure determined by NMR spectroscopy of the pBP136 KlcA protein. The structure shows a novel protein fold and it is clearly not a DNA structural mimic.
- EaStChem School of Chemistry, University of Edinburgh, The King's Buildings, Edinburgh, EH9 3JJ, UK.
Organizational Affiliation: