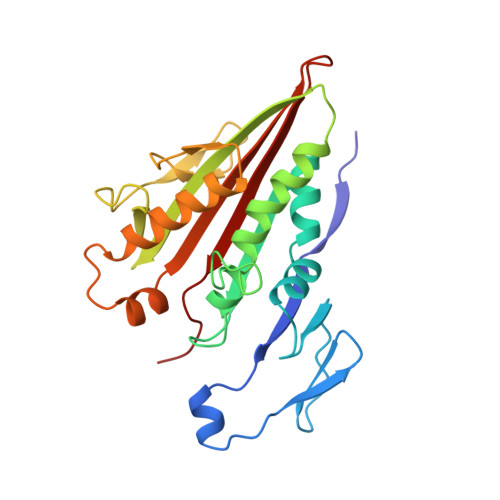
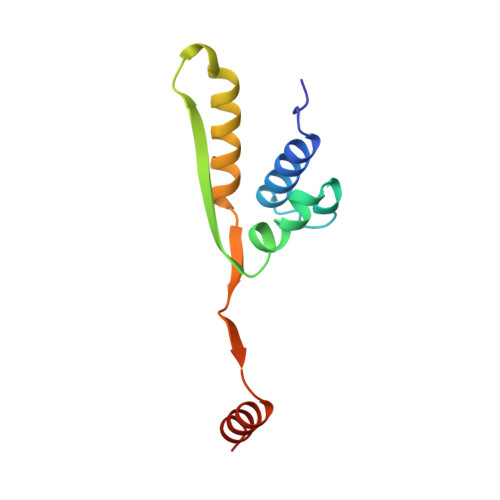
X-ray structure of motor and neck domains from rat brain kinesin.
Sack, S., Muller, J., Marx, A., Thormahlen, M., Mandelkow, E.M., Brady, S.T., Mandelkow, E.(1997) Biochemistry 36: 16155-16165
- PubMed: 9405049
- DOI: https://doi.org/10.1021/bi9722498
- Primary Citation of Related Structures:
2KIN - PubMed Abstract:
We have determined the X-ray structure of rat kinesin head and neck domains. The folding of the core motor domain resembles that of human kinesin reported recently [Kull, F. J., et al. (1996) Nature 380, 550-554]. Novel features of the structure include the N-terminal region, folded as a beta-strand, and the C-terminal transition from the motor to the rod domain, folded as two beta-strands plus an alpha-helix. This helix is the beginning of kinesin's neck responsible for dimerization of the motor complex and for force transduction. Although the folding of the motor domain core is similar to that of a domain of myosin (an actin-dependent motor), the position and angle of kinesin's neck are very different from those of myosin's stalk, suggesting that the two motors have different mechanisms of force transduction. The N- and C-terminal ends of the core motor, thought to be responsible for the directionality of the motors [Case, R. B., et al. (1997) Cell 90, 959-966], take the form of beta-strands attached to the central beta-sheet of the structure.
- Max-Planck-Unit for Structural Molecular Biology, c/o DESY, Notkestrasse 85, D-22607 Hamburg, Germany.
Organizational Affiliation: