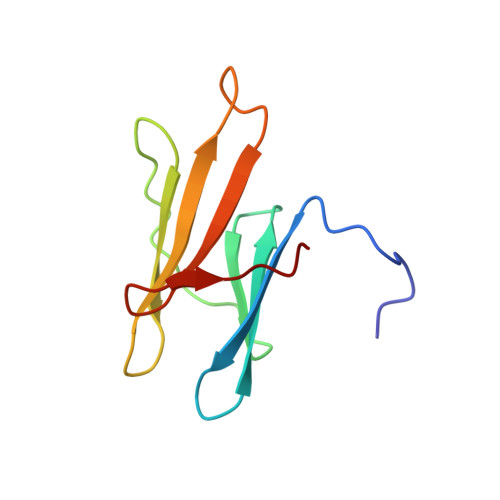
Fas apoptosis inhibitory molecule contains a novel beta-sandwich in contact with a partially ordered domain.
Hemond, M., Rothstein, T.L., Wagner, G.(2009) J Mol Biology 386: 1024-1037
- PubMed: 19168072
- DOI: https://doi.org/10.1016/j.jmb.2009.01.004
- Primary Citation of Related Structures:
2KD2 - PubMed Abstract:
Fas apoptosis inhibitory molecule (FAIM) is a soluble cytosolic protein inhibitor of programmed cell death and is found in organisms throughout the animal kingdom. A short isoform of FAIM is expressed in all tissue types, while an alternatively spliced long isoform is specifically expressed in the brain. Here, the short isoform is shown to consist of two independently folding domains in contact with each other. The NMR solution structure of the C-terminal domain of murine FAIM is solved in isolation and revealed to be a novel protein fold, a noninterleaved seven-stranded beta-sandwich. The structure and sequence reveal several residues that are likely to be involved in functionally significant interactions with the N-terminal domain or other binding partners. Chemical shift perturbation is used to elucidate contacts made between the N-terminal domain and the C-terminal domain.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA.
Organizational Affiliation: