Structural basis of target recognition by ATG8/LC3 during selective autophagy
Noda, N., Kumeta, H., Nakatogawa, H., Adachi, K., Adachi, W., Ishii, J., Fujioka, Y., Ohsumi, Y., Inagaki, F.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Microtubule-associated proteins 1A/1B light chain 3B | 121 | Rattus norvegicus | Mutation(s): 0 Gene Names: Map1lc3b, Map1alc3, Map1lc3 | 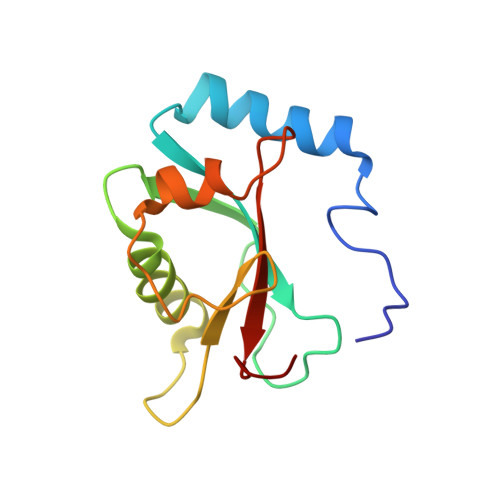 | |
UniProt | |||||
Find proteins for Q62625 (Rattus norvegicus) Explore Q62625 Go to UniProtKB: Q62625 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q62625 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| p62_peptide from Sequestosome-1 | 17 | Rattus norvegicus | Mutation(s): 0 | 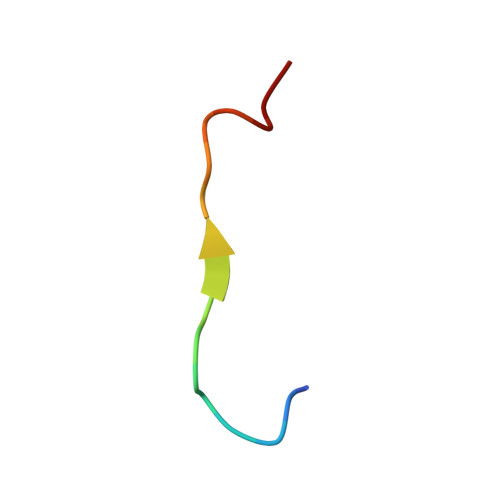 | |
UniProt | |||||
Find proteins for O08623 (Rattus norvegicus) Explore O08623 Go to UniProtKB: O08623 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O08623 | ||||
Sequence AnnotationsExpand | |||||
| |||||